Stergios Christodoulidis
OCTOPUS: Enhancing the Spatial-Awareness of Vision SSMs with Multi-Dimensional Scans and Traversal Selection
Jan 31, 2026Abstract:State space models (SSMs) have recently emerged as an alternative to transformers due to their unique ability of modeling global relationships in text with linear complexity. However, their success in vision tasks has been limited due to their causal formulation, which is suitable for sequential text but detrimental in the spatial domain where causality breaks the inherent spatial relationships among pixels or patches. As a result, standard SSMs fail to capture local spatial coherence, often linking non-adjacent patches while ignoring neighboring ones that are visually correlated. To address these limitations, we introduce OCTOPUS , a novel architecture that preserves both global context and local spatial structure within images, while maintaining the linear complexity of SSMs. OCTOPUS performs discrete reoccurrence along eight principal orientations, going forward or backward in the horizontal, vertical, and diagonal directions, allowing effective information exchange across all spatially connected regions while maintaining independence among unrelated patches. This design enables multi-directional recurrence, capturing both global context and local spatial structure with SSM-level efficiency. In our classification and segmentation benchmarks, OCTOPUS demonstrates notable improvements in boundary preservation and region consistency, as evident from the segmentation results, while maintaining relatively better classification accuracy compared to existing V-SSM based models. These results suggest that OCTOPUS appears as a foundation method for multi-directional recurrence as a scalable and effective mechanism for building spatially aware and computationally efficient vision architectures.
Class Adaptive Conformal Training
Jan 14, 2026Abstract:Deep neural networks have achieved remarkable success across a variety of tasks, yet they often suffer from unreliable probability estimates. As a result, they can be overconfident in their predictions. Conformal Prediction (CP) offers a principled framework for uncertainty quantification, yielding prediction sets with rigorous coverage guarantees. Existing conformal training methods optimize for overall set size, but shaping the prediction sets in a class-conditional manner is not straightforward and typically requires prior knowledge of the data distribution. In this work, we introduce Class Adaptive Conformal Training (CaCT), which formulates conformal training as an augmented Lagrangian optimization problem that adaptively learns to shape prediction sets class-conditionally without making any distributional assumptions. Experiments on multiple benchmark datasets, including standard and long-tailed image recognition as well as text classification, demonstrate that CaCT consistently outperforms prior conformal training methods, producing significantly smaller and more informative prediction sets while maintaining the desired coverage guarantees.
SoC: Semantic Orthogonal Calibration for Test-Time Prompt Tuning
Jan 13, 2026Abstract:With the increasing adoption of vision-language models (VLMs) in critical decision-making systems such as healthcare or autonomous driving, the calibration of their uncertainty estimates becomes paramount. Yet, this dimension has been largely underexplored in the VLM test-time prompt-tuning (TPT) literature, which has predominantly focused on improving their discriminative performance. Recent state-of-the-art advocates for enforcing full orthogonality over pairs of text prompt embeddings to enhance separability, and therefore calibration. Nevertheless, as we theoretically show in this work, the inherent gradients from fully orthogonal constraints will strongly push semantically related classes away, ultimately making the model overconfident. Based on our findings, we propose Semantic Orthogonal Calibration (SoC), a Huber-based regularizer that enforces smooth prototype separation while preserving semantic proximity, thereby improving calibration compared to prior orthogonality-based approaches. Across a comprehensive empirical validation, we demonstrate that SoC consistently improves calibration performance, while also maintaining competitive discriminative capabilities.
CAPRMIL: Context-Aware Patch Representations for Multiple Instance Learning
Dec 16, 2025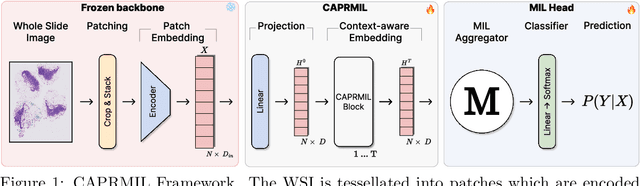
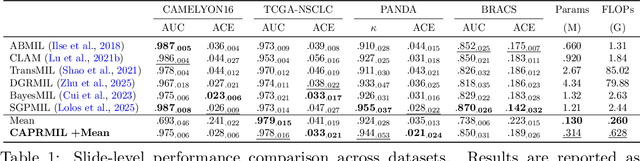
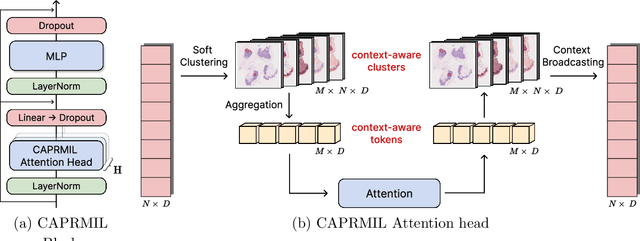
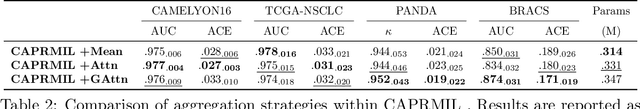
Abstract:In computational pathology, weak supervision has become the standard for deep learning due to the gigapixel scale of WSIs and the scarcity of pixel-level annotations, with Multiple Instance Learning (MIL) established as the principal framework for slide-level model training. In this paper, we introduce a novel setting for MIL methods, inspired by proceedings in Neural Partial Differential Equation (PDE) Solvers. Instead of relying on complex attention-based aggregation, we propose an efficient, aggregator-agnostic framework that removes the complexity of correlation learning from the MIL aggregator. CAPRMIL produces rich context-aware patch embeddings that promote effective correlation learning on downstream tasks. By projecting patch features -- extracted using a frozen patch encoder -- into a small set of global context/morphology-aware tokens and utilizing multi-head self-attention, CAPRMIL injects global context with linear computational complexity with respect to the bag size. Paired with a simple Mean MIL aggregator, CAPRMIL matches state-of-the-art slide-level performance across multiple public pathology benchmarks, while reducing the total number of trainable parameters by 48%-92.8% versus SOTA MILs, lowering FLOPs during inference by 52%-99%, and ranking among the best models on GPU memory efficiency and training time. Our results indicate that learning rich, context-aware instance representations before aggregation is an effective and scalable alternative to complex pooling for whole-slide analysis. Our code is available at https://github.com/mandlos/CAPRMIL
PVeRA: Probabilistic Vector-Based Random Matrix Adaptation
Dec 08, 2025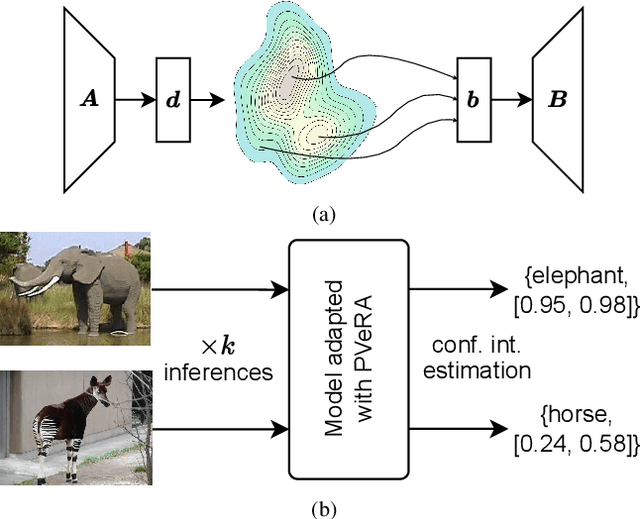
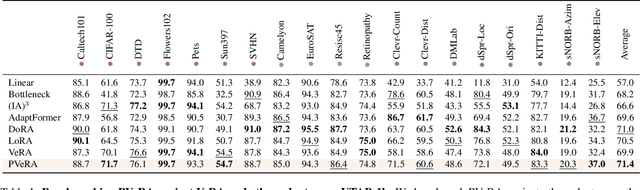
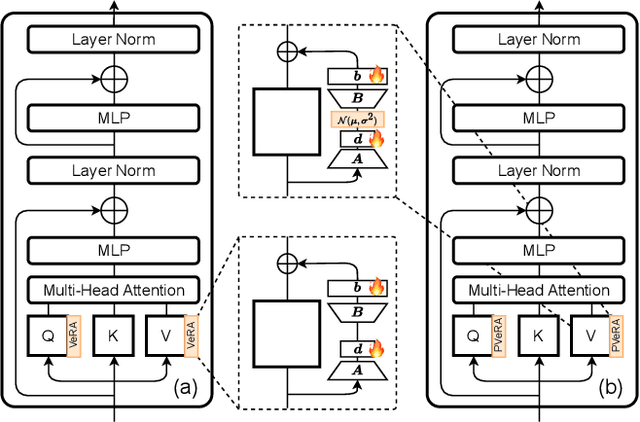

Abstract:Large foundation models have emerged in the last years and are pushing performance boundaries for a variety of tasks. Training or even finetuning such models demands vast datasets and computational resources, which are often scarce and costly. Adaptation methods provide a computationally efficient solution to address these limitations by allowing such models to be finetuned on small amounts of data and computing power. This is achieved by appending new trainable modules to frozen backbones with only a fraction of the trainable parameters and fitting only these modules on novel tasks. Recently, the VeRA adapter was shown to excel in parameter-efficient adaptations by utilizing a pair of frozen random low-rank matrices shared across all layers. In this paper, we propose PVeRA, a probabilistic version of the VeRA adapter, which modifies the low-rank matrices of VeRA in a probabilistic manner. This modification naturally allows handling inherent ambiguities in the input and allows for different sampling configurations during training and testing. A comprehensive evaluation was performed on the VTAB-1k benchmark and seven adapters, with PVeRA outperforming VeRA and other adapters. Our code for training models with PVeRA and benchmarking all adapters is available https://github.com/leofillioux/pvera.
Controllable Latent Space Augmentation for Digital Pathology
Aug 20, 2025Abstract:Whole slide image (WSI) analysis in digital pathology presents unique challenges due to the gigapixel resolution of WSIs and the scarcity of dense supervision signals. While Multiple Instance Learning (MIL) is a natural fit for slide-level tasks, training robust models requires large and diverse datasets. Even though image augmentation techniques could be utilized to increase data variability and reduce overfitting, implementing them effectively is not a trivial task. Traditional patch-level augmentation is prohibitively expensive due to the large number of patches extracted from each WSI, and existing feature-level augmentation methods lack control over transformation semantics. We introduce HistAug, a fast and efficient generative model for controllable augmentations in the latent space for digital pathology. By conditioning on explicit patch-level transformations (e.g., hue, erosion), HistAug generates realistic augmented embeddings while preserving initial semantic information. Our method allows the processing of a large number of patches in a single forward pass efficiently, while at the same time consistently improving MIL model performance. Experiments across multiple slide-level tasks and diverse organs show that HistAug outperforms existing methods, particularly in low-data regimes. Ablation studies confirm the benefits of learned transformations over noise-based perturbations and highlight the importance of uniform WSI-wise augmentation. Code is available at https://github.com/MICS-Lab/HistAug.
On the Risk of Misleading Reports: Diagnosing Textual Biases in Multimodal Clinical AI
Jul 31, 2025



Abstract:Clinical decision-making relies on the integrated analysis of medical images and the associated clinical reports. While Vision-Language Models (VLMs) can offer a unified framework for such tasks, they can exhibit strong biases toward one modality, frequently overlooking critical visual cues in favor of textual information. In this work, we introduce Selective Modality Shifting (SMS), a perturbation-based approach to quantify a model's reliance on each modality in binary classification tasks. By systematically swapping images or text between samples with opposing labels, we expose modality-specific biases. We assess six open-source VLMs-four generalist models and two fine-tuned for medical data-on two medical imaging datasets with distinct modalities: MIMIC-CXR (chest X-ray) and FairVLMed (scanning laser ophthalmoscopy). By assessing model performance and the calibration of every model in both unperturbed and perturbed settings, we reveal a marked dependency on text input, which persists despite the presence of complementary visual information. We also perform a qualitative attention-based analysis which further confirms that image content is often overshadowed by text details. Our findings highlight the importance of designing and evaluating multimodal medical models that genuinely integrate visual and textual cues, rather than relying on single-modality signals.
THUNDER: Tile-level Histopathology image UNDERstanding benchmark
Jul 10, 2025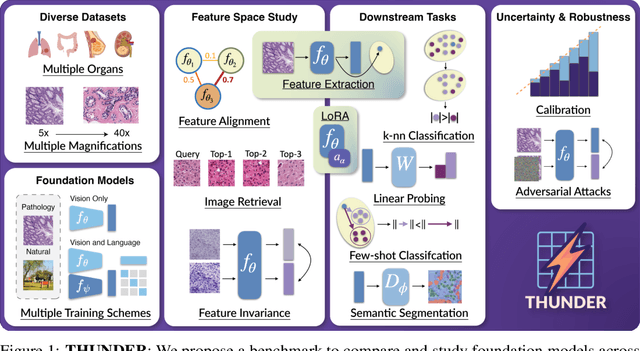
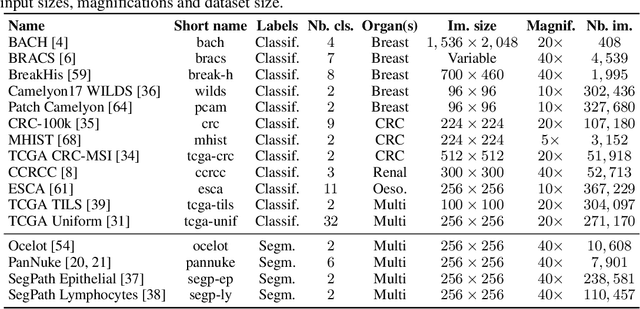

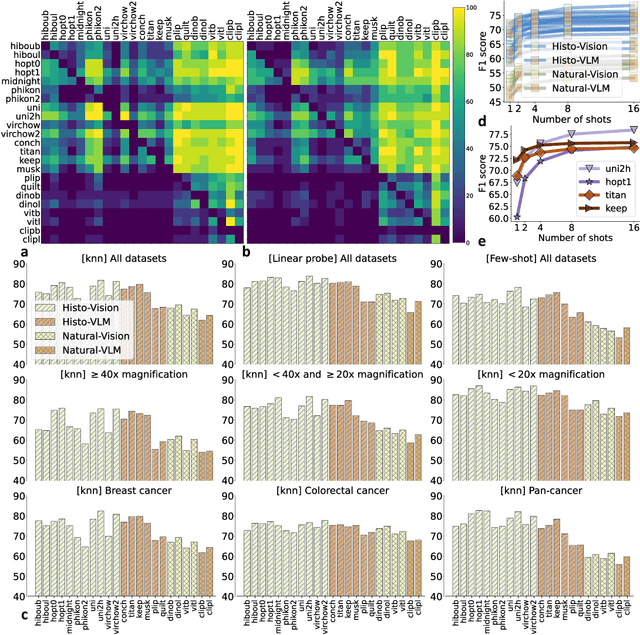
Abstract:Progress in a research field can be hard to assess, in particular when many concurrent methods are proposed in a short period of time. This is the case in digital pathology, where many foundation models have been released recently to serve as feature extractors for tile-level images, being used in a variety of downstream tasks, both for tile- and slide-level problems. Benchmarking available methods then becomes paramount to get a clearer view of the research landscape. In particular, in critical domains such as healthcare, a benchmark should not only focus on evaluating downstream performance, but also provide insights about the main differences between methods, and importantly, further consider uncertainty and robustness to ensure a reliable usage of proposed models. For these reasons, we introduce THUNDER, a tile-level benchmark for digital pathology foundation models, allowing for efficient comparison of many models on diverse datasets with a series of downstream tasks, studying their feature spaces and assessing the robustness and uncertainty of predictions informed by their embeddings. THUNDER is a fast, easy-to-use, dynamic benchmark that can already support a large variety of state-of-the-art foundation, as well as local user-defined models for direct tile-based comparison. In this paper, we provide a comprehensive comparison of 23 foundation models on 16 different datasets covering diverse tasks, feature analysis, and robustness. The code for THUNDER is publicly available at https://github.com/MICS-Lab/thunder.
Full Conformal Adaptation of Medical Vision-Language Models
Jun 06, 2025



Abstract:Vision-language models (VLMs) pre-trained at large scale have shown unprecedented transferability capabilities and are being progressively integrated into medical image analysis. Although its discriminative potential has been widely explored, its reliability aspect remains overlooked. This work investigates their behavior under the increasingly popular split conformal prediction (SCP) framework, which theoretically guarantees a given error level on output sets by leveraging a labeled calibration set. However, the zero-shot performance of VLMs is inherently limited, and common practice involves few-shot transfer learning pipelines, which cannot absorb the rigid exchangeability assumptions of SCP. To alleviate this issue, we propose full conformal adaptation, a novel setting for jointly adapting and conformalizing pre-trained foundation models, which operates transductively over each test data point using a few-shot adaptation set. Moreover, we complement this framework with SS-Text, a novel training-free linear probe solver for VLMs that alleviates the computational cost of such a transductive approach. We provide comprehensive experiments using 3 different modality-specialized medical VLMs and 9 adaptation tasks. Our framework requires exactly the same data as SCP, and provides consistent relative improvements of up to 27% on set efficiency while maintaining the same coverage guarantees.
BayesAdapter: enhanced uncertainty estimation in CLIP few-shot adaptation
Dec 12, 2024



Abstract:The emergence of large pre-trained vision-language models (VLMs) represents a paradigm shift in machine learning, with unprecedented results in a broad span of visual recognition tasks. CLIP, one of the most popular VLMs, has exhibited remarkable zero-shot and transfer learning capabilities in classification. To transfer CLIP to downstream tasks, adapters constitute a parameter-efficient approach that avoids backpropagation through the large model (unlike related prompt learning methods). However, CLIP adapters have been developed to target discriminative performance, and the quality of their uncertainty estimates has been overlooked. In this work we show that the discriminative performance of state-of-the-art CLIP adapters does not always correlate with their uncertainty estimation capabilities, which are essential for a safe deployment in real-world scenarios. We also demonstrate that one of such adapters is obtained through MAP inference from a more general probabilistic framework. Based on this observation we introduce BayesAdapter, which leverages Bayesian inference to estimate a full probability distribution instead of a single point, better capturing the variability inherent in the parameter space. In a comprehensive empirical evaluation we show that our approach obtains high quality uncertainty estimates in the predictions, standing out in calibration and selective classification. Our code is publicly available at: https://github.com/pablomorales92/BayesAdapter.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge