Leo Fillioux
SoC: Semantic Orthogonal Calibration for Test-Time Prompt Tuning
Jan 13, 2026Abstract:With the increasing adoption of vision-language models (VLMs) in critical decision-making systems such as healthcare or autonomous driving, the calibration of their uncertainty estimates becomes paramount. Yet, this dimension has been largely underexplored in the VLM test-time prompt-tuning (TPT) literature, which has predominantly focused on improving their discriminative performance. Recent state-of-the-art advocates for enforcing full orthogonality over pairs of text prompt embeddings to enhance separability, and therefore calibration. Nevertheless, as we theoretically show in this work, the inherent gradients from fully orthogonal constraints will strongly push semantically related classes away, ultimately making the model overconfident. Based on our findings, we propose Semantic Orthogonal Calibration (SoC), a Huber-based regularizer that enforces smooth prototype separation while preserving semantic proximity, thereby improving calibration compared to prior orthogonality-based approaches. Across a comprehensive empirical validation, we demonstrate that SoC consistently improves calibration performance, while also maintaining competitive discriminative capabilities.
PVeRA: Probabilistic Vector-Based Random Matrix Adaptation
Dec 08, 2025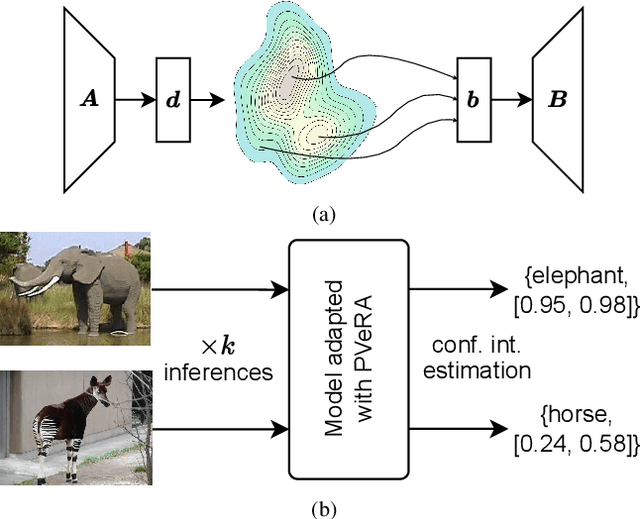
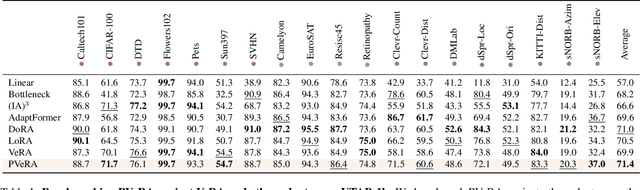
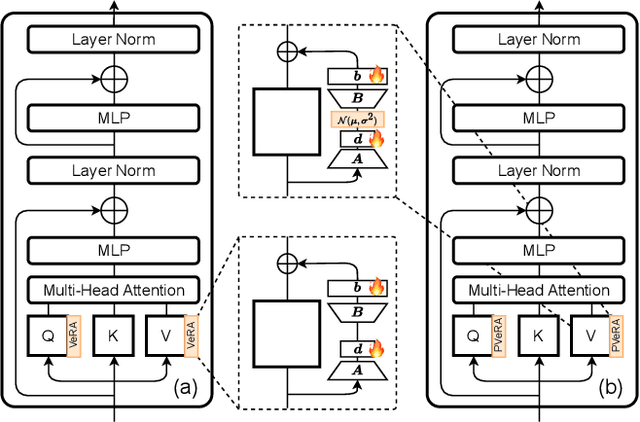

Abstract:Large foundation models have emerged in the last years and are pushing performance boundaries for a variety of tasks. Training or even finetuning such models demands vast datasets and computational resources, which are often scarce and costly. Adaptation methods provide a computationally efficient solution to address these limitations by allowing such models to be finetuned on small amounts of data and computing power. This is achieved by appending new trainable modules to frozen backbones with only a fraction of the trainable parameters and fitting only these modules on novel tasks. Recently, the VeRA adapter was shown to excel in parameter-efficient adaptations by utilizing a pair of frozen random low-rank matrices shared across all layers. In this paper, we propose PVeRA, a probabilistic version of the VeRA adapter, which modifies the low-rank matrices of VeRA in a probabilistic manner. This modification naturally allows handling inherent ambiguities in the input and allows for different sampling configurations during training and testing. A comprehensive evaluation was performed on the VTAB-1k benchmark and seven adapters, with PVeRA outperforming VeRA and other adapters. Our code for training models with PVeRA and benchmarking all adapters is available https://github.com/leofillioux/pvera.
THUNDER: Tile-level Histopathology image UNDERstanding benchmark
Jul 10, 2025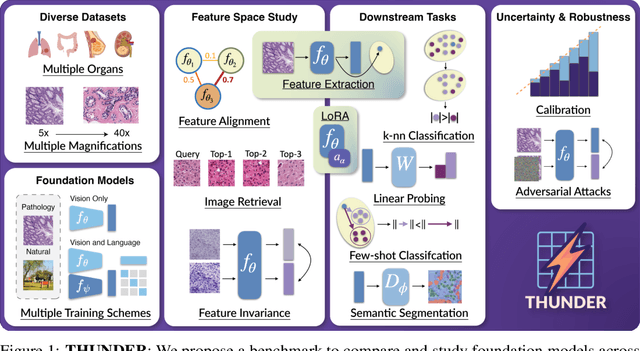
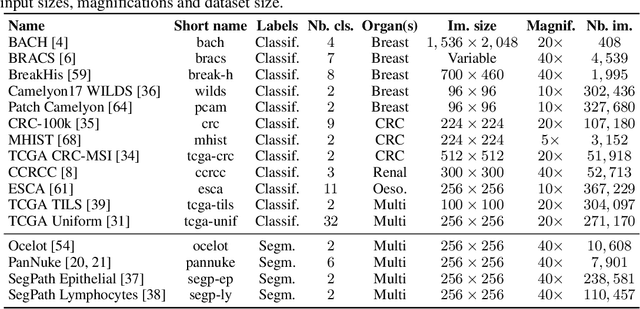

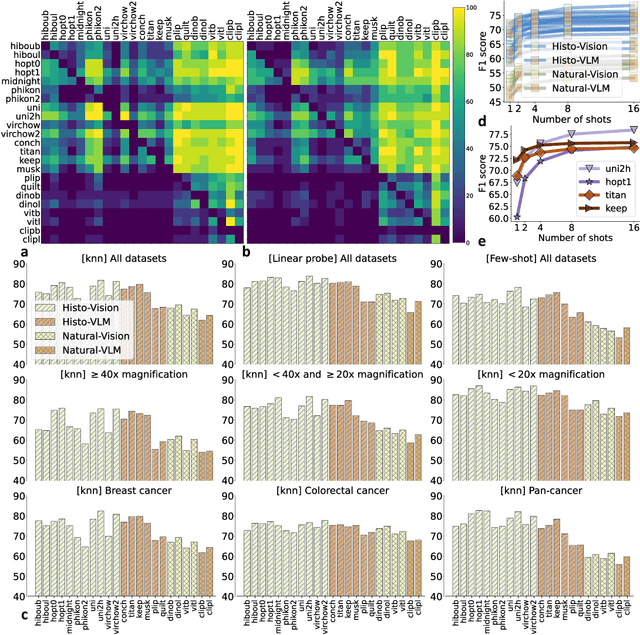
Abstract:Progress in a research field can be hard to assess, in particular when many concurrent methods are proposed in a short period of time. This is the case in digital pathology, where many foundation models have been released recently to serve as feature extractors for tile-level images, being used in a variety of downstream tasks, both for tile- and slide-level problems. Benchmarking available methods then becomes paramount to get a clearer view of the research landscape. In particular, in critical domains such as healthcare, a benchmark should not only focus on evaluating downstream performance, but also provide insights about the main differences between methods, and importantly, further consider uncertainty and robustness to ensure a reliable usage of proposed models. For these reasons, we introduce THUNDER, a tile-level benchmark for digital pathology foundation models, allowing for efficient comparison of many models on diverse datasets with a series of downstream tasks, studying their feature spaces and assessing the robustness and uncertainty of predictions informed by their embeddings. THUNDER is a fast, easy-to-use, dynamic benchmark that can already support a large variety of state-of-the-art foundation, as well as local user-defined models for direct tile-based comparison. In this paper, we provide a comprehensive comparison of 23 foundation models on 16 different datasets covering diverse tasks, feature analysis, and robustness. The code for THUNDER is publicly available at https://github.com/MICS-Lab/thunder.
Full Conformal Adaptation of Medical Vision-Language Models
Jun 06, 2025



Abstract:Vision-language models (VLMs) pre-trained at large scale have shown unprecedented transferability capabilities and are being progressively integrated into medical image analysis. Although its discriminative potential has been widely explored, its reliability aspect remains overlooked. This work investigates their behavior under the increasingly popular split conformal prediction (SCP) framework, which theoretically guarantees a given error level on output sets by leveraging a labeled calibration set. However, the zero-shot performance of VLMs is inherently limited, and common practice involves few-shot transfer learning pipelines, which cannot absorb the rigid exchangeability assumptions of SCP. To alleviate this issue, we propose full conformal adaptation, a novel setting for jointly adapting and conformalizing pre-trained foundation models, which operates transductively over each test data point using a few-shot adaptation set. Moreover, we complement this framework with SS-Text, a novel training-free linear probe solver for VLMs that alleviates the computational cost of such a transductive approach. We provide comprehensive experiments using 3 different modality-specialized medical VLMs and 9 adaptation tasks. Our framework requires exactly the same data as SCP, and provides consistent relative improvements of up to 27% on set efficiency while maintaining the same coverage guarantees.
Are foundation models for computer vision good conformal predictors?
Dec 08, 2024



Abstract:Recent advances in self-supervision and constrastive learning have brought the performance of foundation models to unprecedented levels in a variety of tasks. Fueled by this progress, these models are becoming the prevailing approach for a wide array of real-world vision problems, including risk-sensitive and high-stakes applications. However, ensuring safe deployment in these scenarios requires a more comprehensive understanding of their uncertainty modeling capabilities, which has been barely explored. In this work, we delve into the behavior of vision and vision-language foundation models under Conformal Prediction (CP), a statistical framework that provides theoretical guarantees of marginal coverage of the true class. Across extensive experiments including popular vision classification benchmarks, well-known foundation vision models, and three CP methods, our findings reveal that foundation models are well-suited for conformalization procedures, particularly those integrating Vision Transformers. Furthermore, we show that calibrating the confidence predictions of these models leads to efficiency degradation of the conformal set on adaptive CP methods. In contrast, few-shot adaptation to downstream tasks generally enhances conformal scores, where we identify Adapters as a better conformable alternative compared to Prompt Learning strategies. Our empirical study identifies APS as particularly promising in the context of vision foundation models, as it does not violate the marginal coverage property across multiple challenging, yet realistic scenarios.
Spatio-Temporal Analysis of Patient-Derived Organoid Videos Using Deep Learning for the Prediction of Drug Efficacy
Aug 28, 2023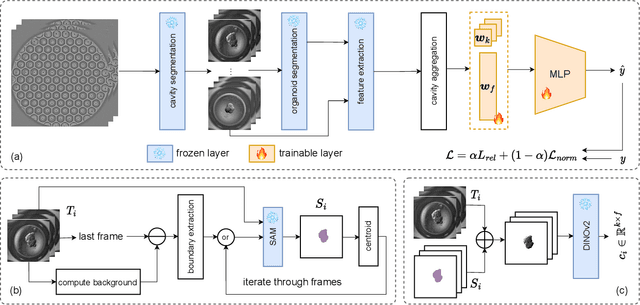

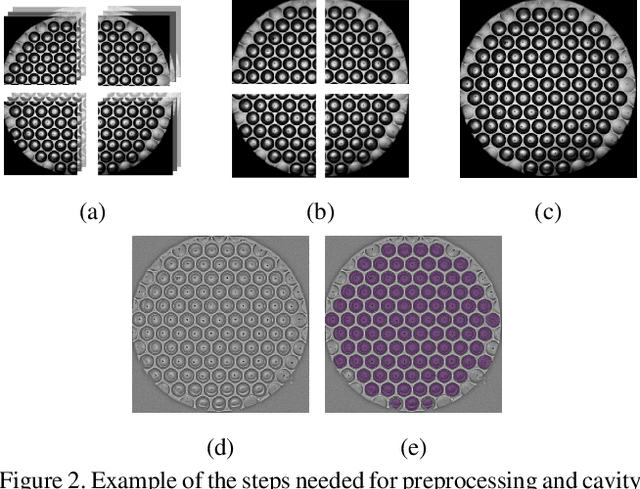

Abstract:Over the last ten years, Patient-Derived Organoids (PDOs) emerged as the most reliable technology to generate ex-vivo tumor avatars. PDOs retain the main characteristics of their original tumor, making them a system of choice for pre-clinical and clinical studies. In particular, PDOs are attracting interest in the field of Functional Precision Medicine (FPM), which is based upon an ex-vivo drug test in which living tumor cells (such as PDOs) from a specific patient are exposed to a panel of anti-cancer drugs. Currently, the Adenosine Triphosphate (ATP) based cell viability assay is the gold standard test to assess the sensitivity of PDOs to drugs. The readout is measured at the end of the assay from a global PDO population and therefore does not capture single PDO responses and does not provide time resolution of drug effect. To this end, in this study, we explore for the first time the use of powerful large foundation models for the automatic processing of PDO data. In particular, we propose a novel imaging-based high-throughput screening method to assess real-time drug efficacy from a time-lapse microscopy video of PDOs. The recently proposed SAM algorithm for segmentation and DINOv2 model are adapted in a comprehensive pipeline for processing PDO microscopy frames. Moreover, an attention mechanism is proposed for fusing temporal and spatial features in a multiple instance learning setting to predict ATP. We report better results than other non-time-resolved methods, indicating that the temporality of data is an important factor for the prediction of ATP. Extensive ablations shed light on optimizing the experimental setting and automating the prediction both in real-time and for forecasting.
Structured State Space Models for Multiple Instance Learning in Digital Pathology
Jun 27, 2023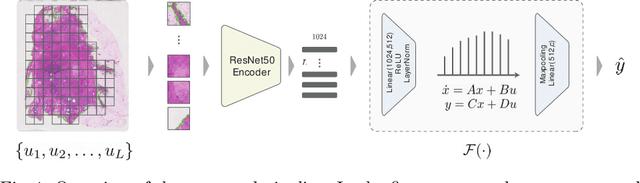
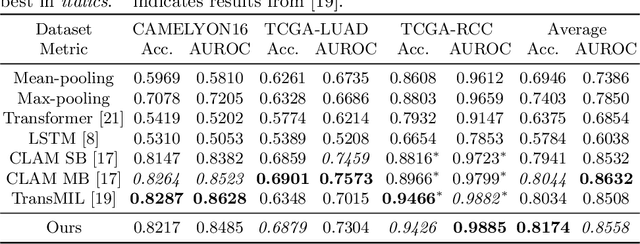

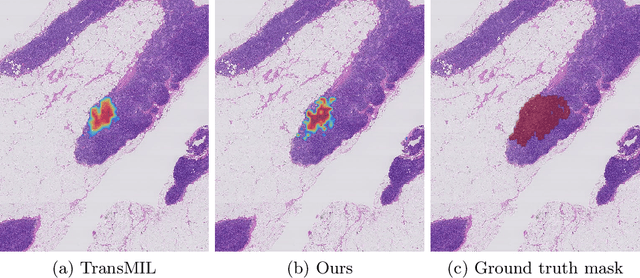
Abstract:Multiple instance learning is an ideal mode of analysis for histopathology data, where vast whole slide images are typically annotated with a single global label. In such cases, a whole slide image is modelled as a collection of tissue patches to be aggregated and classified. Common models for performing this classification include recurrent neural networks and transformers. Although powerful compression algorithms, such as deep pre-trained neural networks, are used to reduce the dimensionality of each patch, the sequences arising from whole slide images remain excessively long, routinely containing tens of thousands of patches. Structured state space models are an emerging alternative for sequence modelling, specifically designed for the efficient modelling of long sequences. These models invoke an optimal projection of an input sequence into memory units that compress the entire sequence. In this paper, we propose the use of state space models as a multiple instance learner to a variety of problems in digital pathology. Across experiments in metastasis detection, cancer subtyping, mutation classification, and multitask learning, we demonstrate the competitiveness of this new class of models with existing state of the art approaches. Our code is available at https://github.com/MICS-Lab/s4_digital_pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge