Sabrina Bedja
Spatio-Temporal Analysis of Patient-Derived Organoid Videos Using Deep Learning for the Prediction of Drug Efficacy
Aug 28, 2023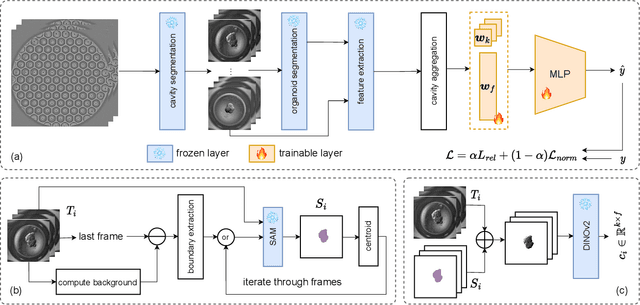

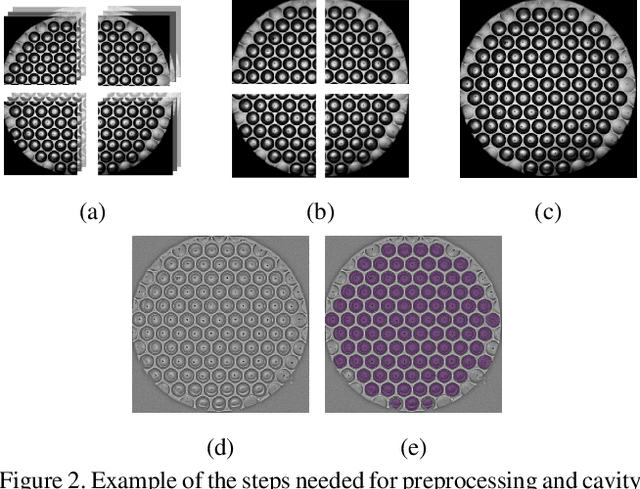

Abstract:Over the last ten years, Patient-Derived Organoids (PDOs) emerged as the most reliable technology to generate ex-vivo tumor avatars. PDOs retain the main characteristics of their original tumor, making them a system of choice for pre-clinical and clinical studies. In particular, PDOs are attracting interest in the field of Functional Precision Medicine (FPM), which is based upon an ex-vivo drug test in which living tumor cells (such as PDOs) from a specific patient are exposed to a panel of anti-cancer drugs. Currently, the Adenosine Triphosphate (ATP) based cell viability assay is the gold standard test to assess the sensitivity of PDOs to drugs. The readout is measured at the end of the assay from a global PDO population and therefore does not capture single PDO responses and does not provide time resolution of drug effect. To this end, in this study, we explore for the first time the use of powerful large foundation models for the automatic processing of PDO data. In particular, we propose a novel imaging-based high-throughput screening method to assess real-time drug efficacy from a time-lapse microscopy video of PDOs. The recently proposed SAM algorithm for segmentation and DINOv2 model are adapted in a comprehensive pipeline for processing PDO microscopy frames. Moreover, an attention mechanism is proposed for fusing temporal and spatial features in a multiple instance learning setting to predict ATP. We report better results than other non-time-resolved methods, indicating that the temporality of data is an important factor for the prediction of ATP. Extensive ablations shed light on optimizing the experimental setting and automating the prediction both in real-time and for forecasting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge