Fabrice André
On the detection of Out-Of-Distribution samples in Multiple Instance Learning
Sep 11, 2023

Abstract:The deployment of machine learning solutions in real-world scenarios often involves addressing the challenge of out-of-distribution (OOD) detection. While significant efforts have been devoted to OOD detection in classical supervised settings, the context of weakly supervised learning, particularly the Multiple Instance Learning (MIL) framework, remains under-explored. In this study, we tackle this challenge by adapting post-hoc OOD detection methods to the MIL setting while introducing a novel benchmark specifically designed to assess OOD detection performance in weakly supervised scenarios. Extensive experiments based on diverse public datasets do not reveal a single method with a clear advantage over the others. Although DICE emerges as the best-performing method overall, it exhibits significant shortcomings on some datasets, emphasizing the complexity of this under-explored and challenging topic. Our findings shed light on the complex nature of OOD detection under the MIL framework, emphasizing the importance of developing novel, robust, and reliable methods that can generalize effectively in a weakly supervised context. The code for the paper is available here: https://github.com/loic-lb/OOD_MIL.
Interpretable cytometry cell-type annotation with flow-based deep generative models
Aug 11, 2022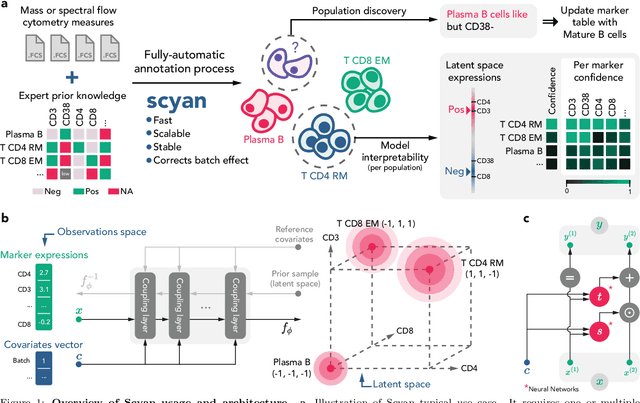
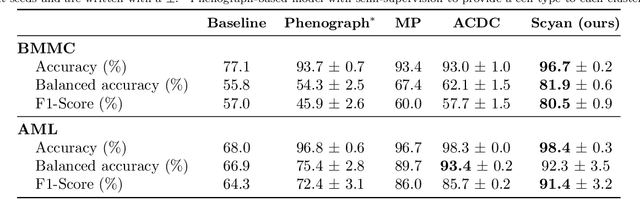
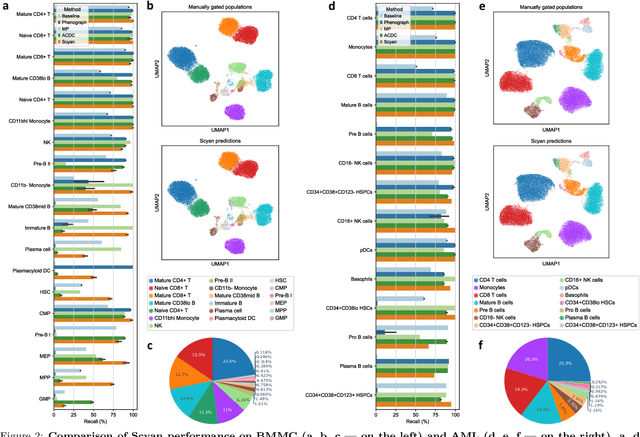
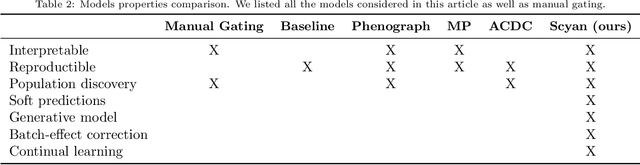
Abstract:Cytometry enables precise single-cell phenotyping within heterogeneous populations. These cell types are traditionally annotated via manual gating, but this method suffers from a lack of reproducibility and sensitivity to batch-effect. Also, the most recent cytometers - spectral flow or mass cytometers - create rich and high-dimensional data whose analysis via manual gating becomes challenging and time-consuming. To tackle these limitations, we introduce Scyan (https://github.com/MICS-Lab/scyan), a Single-cell Cytometry Annotation Network that automatically annotates cell types using only prior expert knowledge about the cytometry panel. We demonstrate that Scyan significantly outperforms the related state-of-the-art models on multiple public datasets while being faster and interpretable. In addition, Scyan overcomes several complementary tasks such as batch-effect removal, debarcoding, and population discovery. Overall, this model accelerates and eases cell population characterisation, quantification, and discovery in cytometry.
Self-Supervised Nuclei Segmentation in Histopathological Images Using Attention
Jul 16, 2020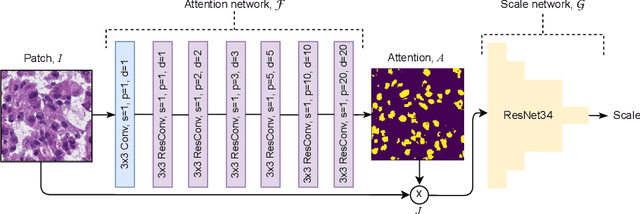
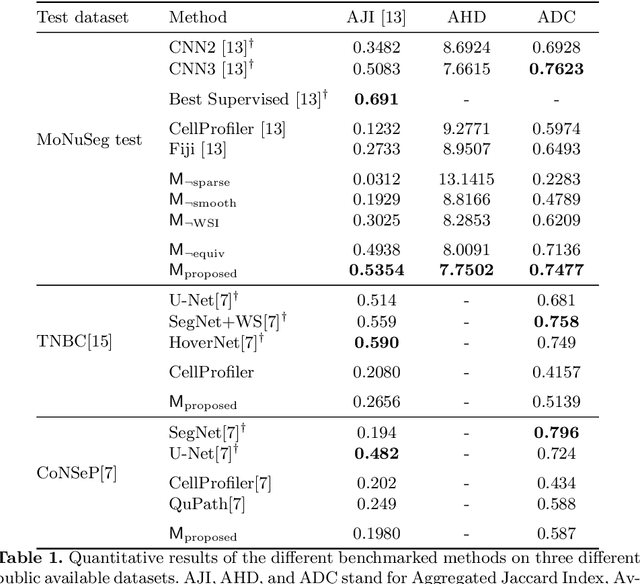
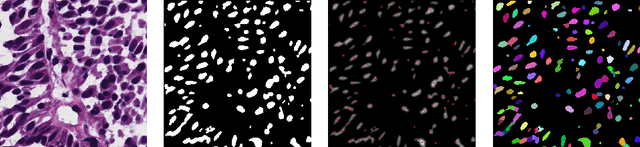
Abstract:Segmentation and accurate localization of nuclei in histopathological images is a very challenging problem, with most existing approaches adopting a supervised strategy. These methods usually rely on manual annotations that require a lot of time and effort from medical experts. In this study, we present a self-supervised approach for segmentation of nuclei for whole slide histopathology images. Our method works on the assumption that the size and texture of nuclei can determine the magnification at which a patch is extracted. We show that the identification of the magnification level for tiles can generate a preliminary self-supervision signal to locate nuclei. We further show that by appropriately constraining our model it is possible to retrieve meaningful segmentation maps as an auxiliary output to the primary magnification identification task. Our experiments show that with standard post-processing, our method can outperform other unsupervised nuclei segmentation approaches and report similar performance with supervised ones on the publicly available MoNuSeg dataset. Our code and models are available online to facilitate further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge