Stefan Michiels
CustOmics: A versatile deep-learning based strategy for multi-omics integration
Sep 12, 2022
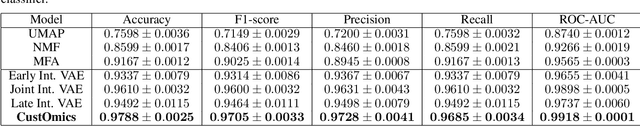
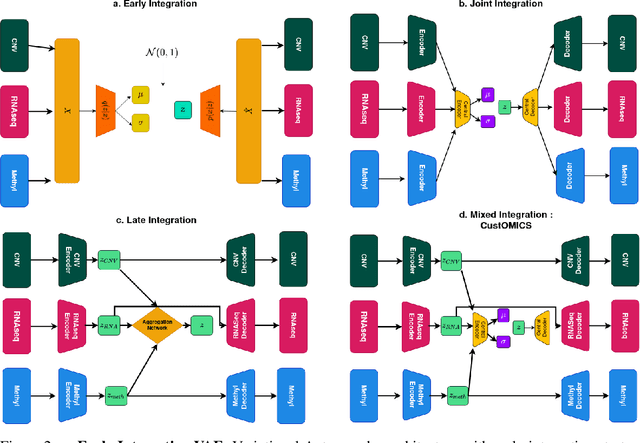

Abstract:Recent advances in high-throughput sequencing technologies have enabled the extraction of multiple features that depict patient samples at diverse and complementary molecular levels. The generation of such data has led to new challenges in computational biology regarding the integration of high-dimensional and heterogeneous datasets that capture the interrelationships between multiple genes and their functions. Thanks to their versatility and ability to learn synthetic latent representations of complex data, deep learning methods offer promising perspectives for integrating multi-omics data. These methods have led to the conception of many original architectures that are primarily based on autoencoder models. However, due to the difficulty of the task, the integration strategy is fundamental to take full advantage of the sources' particularities without losing the global trends. This paper presents a novel strategy to build a customizable autoencoder model that adapts to the dataset used in the case of high-dimensional multi-source integration. We will assess the impact of integration strategies on the latent representation and combine the best strategies to propose a new method, CustOmics (https://github.com/HakimBenkirane/CustOmics). We focus here on the integration of data from multiple omics sources and demonstrate the performance of the proposed method on test cases for several tasks such as classification and survival analysis.
Self-Supervised Nuclei Segmentation in Histopathological Images Using Attention
Jul 16, 2020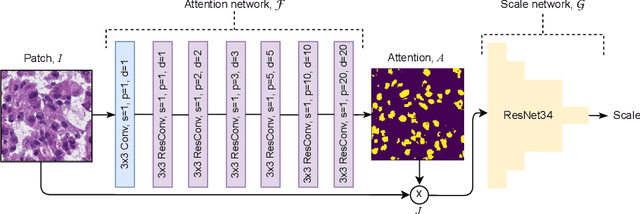
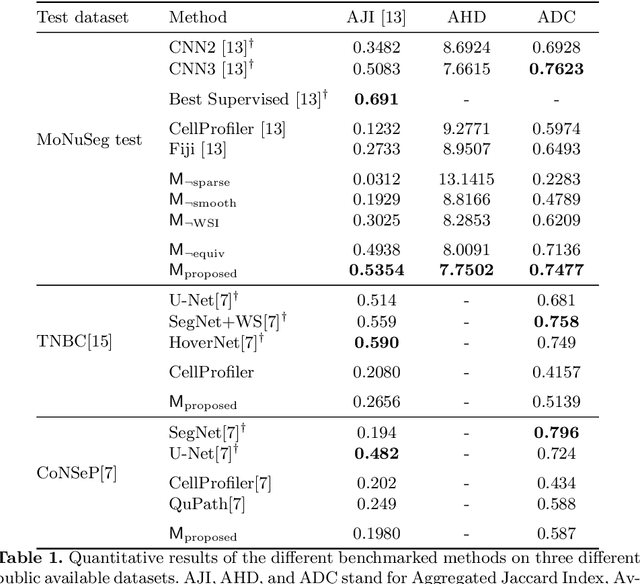
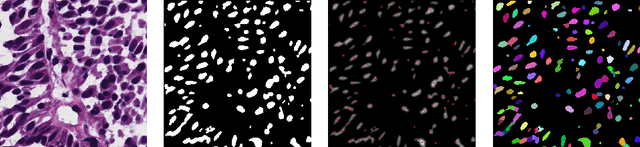
Abstract:Segmentation and accurate localization of nuclei in histopathological images is a very challenging problem, with most existing approaches adopting a supervised strategy. These methods usually rely on manual annotations that require a lot of time and effort from medical experts. In this study, we present a self-supervised approach for segmentation of nuclei for whole slide histopathology images. Our method works on the assumption that the size and texture of nuclei can determine the magnification at which a patch is extracted. We show that the identification of the magnification level for tiles can generate a preliminary self-supervision signal to locate nuclei. We further show that by appropriately constraining our model it is possible to retrieve meaningful segmentation maps as an auxiliary output to the primary magnification identification task. Our experiments show that with standard post-processing, our method can outperform other unsupervised nuclei segmentation approaches and report similar performance with supervised ones on the publicly available MoNuSeg dataset. Our code and models are available online to facilitate further research.
Estimating causal effects of time-dependent exposures on a binary endpoint in a high-dimensional setting
Mar 29, 2018
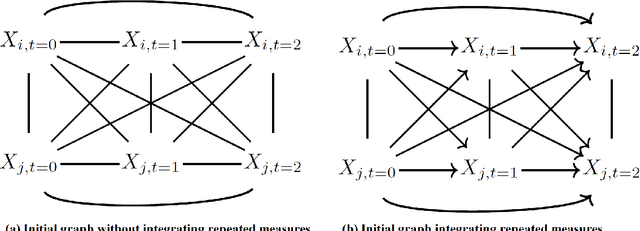
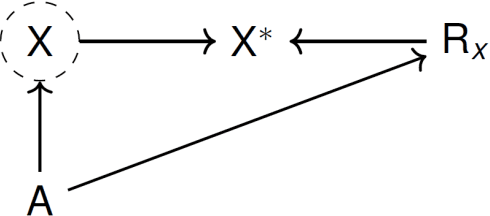
Abstract:Recently, the intervention calculus when the DAG is absent (IDA) method was developed to estimate lower bounds of causal effects from observational high-dimensional data. Originally it was introduced to assess the effect of baseline biomarkers which do not vary over time. However, in many clinical settings, measurements of biomarkers are repeated at fixed time points during treatment exposure and, therefore, this method need to be extended. The purpose of this paper is then to extend the first step of the IDA, the Peter Clarks (PC)-algorithm, to a time-dependent exposure in the context of a binary outcome. We generalised the PC-algorithm for taking into account the chronological order of repeated measurements of the exposure and propose to apply the IDA with our new version, the chronologically ordered PC-algorithm (COPC-algorithm). A simulation study has been performed before applying the method for estimating causal effects of time-dependent immunological biomarkers on toxicity, death and progression in patients with metastatic melanoma. The simulation study showed that the completed partially directed acyclic graphs (CPDAGs) obtained using COPC-algorithm were structurally closer to the true CPDAG than CPDAGs obtained using PC-algorithm. Also, causal effects were more accurate when they were estimated based on CPDAGs obtained using COPC-algorithm. Moreover, CPDAGs obtained by COPC-algorithm allowed removing non-chronologic arrows with a variable measured at a time t pointing to a variable measured at a time t' where t'< t. Bidirected edges were less present in CPDAGs obtained with the COPC-algorithm, supporting the fact that there was less variability in causal effects estimated from these CPDAGs. The COPC-algorithm provided CPDAGs that keep the chronological structure present in the data, thus allowed to estimate lower bounds of the causal effect of time-dependent biomarkers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge