Sunil Kumar Sahu
Bilingual Adaptation of Monolingual Foundation Models
Jul 13, 2024
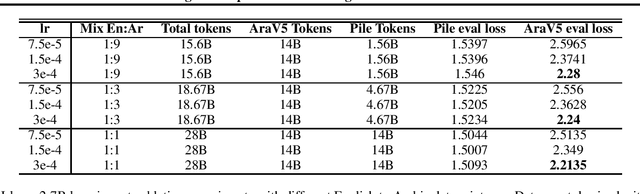


Abstract:We present an efficient method for adapting a monolingual Large Language Model (LLM) to another language, addressing challenges of catastrophic forgetting and tokenizer limitations. We focus this study on adapting Llama 2 to Arabic. Our two-stage approach begins with expanding the vocabulary and training only the embeddings matrix, followed by full model continual pretraining on a bilingual corpus. By continually pretraining on a mix of Arabic and English corpora, the model retains its proficiency in English while acquiring capabilities in Arabic. Our approach results in significant improvements in Arabic and slight enhancements in English, demonstrating cost-effective cross-lingual transfer. We also perform extensive ablations on embedding initialization techniques, data mix ratios, and learning rates and release a detailed training recipe.
Jais and Jais-chat: Arabic-Centric Foundation and Instruction-Tuned Open Generative Large Language Models
Aug 30, 2023
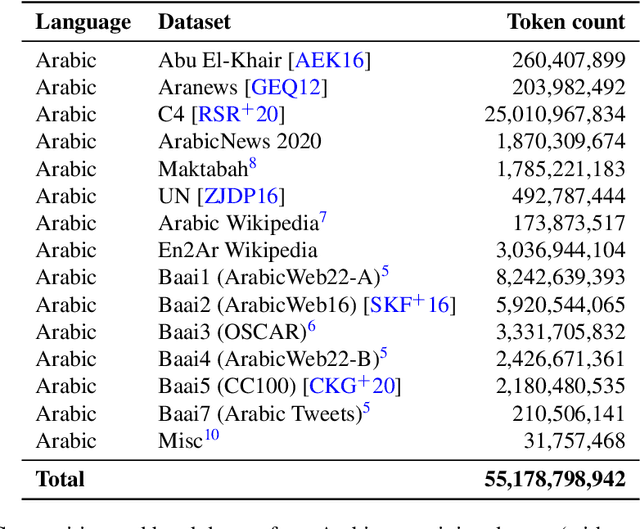
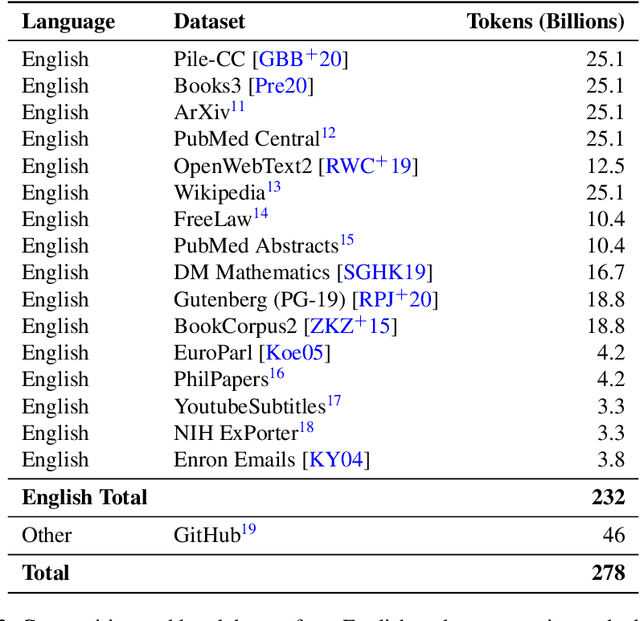
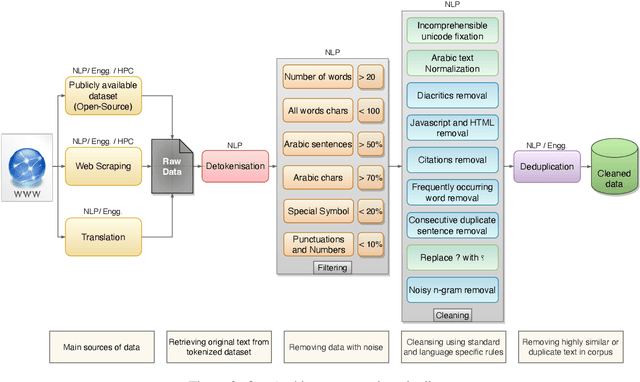
Abstract:We introduce Jais and Jais-chat, new state-of-the-art Arabic-centric foundation and instruction-tuned open generative large language models (LLMs). The models are based on the GPT-3 decoder-only architecture and are pretrained on a mixture of Arabic and English texts, including source code in various programming languages. With 13 billion parameters, they demonstrate better knowledge and reasoning capabilities in Arabic than any existing open Arabic and multilingual models by a sizable margin, based on extensive evaluation. Moreover, the models are competitive in English compared to English-centric open models of similar size, despite being trained on much less English data. We provide a detailed description of the training, the tuning, the safety alignment, and the evaluation of the models. We release two open versions of the model -- the foundation Jais model, and an instruction-tuned Jais-chat variant -- with the aim of promoting research on Arabic LLMs. Available at https://huggingface.co/inception-mbzuai/jais-13b-chat
Relation Extraction with Self-determined Graph Convolutional Network
Aug 27, 2020



Abstract:Relation Extraction is a way of obtaining the semantic relationship between entities in text. The state-of-the-art methods use linguistic tools to build a graph for the text in which the entities appear and then a Graph Convolutional Network (GCN) is employed to encode the pre-built graphs. Although their performance is promising, the reliance on linguistic tools results in a non end-to-end process. In this work, we propose a novel model, the Self-determined Graph Convolutional Network (SGCN), which determines a weighted graph using a self-attention mechanism, rather using any linguistic tool. Then, the self-determined graph is encoded using a GCN. We test our model on the TACRED dataset and achieve the state-of-the-art result. Our experiments show that SGCN outperforms the traditional GCN, which uses dependency parsing tools to build the graph.
Inter-sentence Relation Extraction with Document-level Graph Convolutional Neural Network
Jun 11, 2019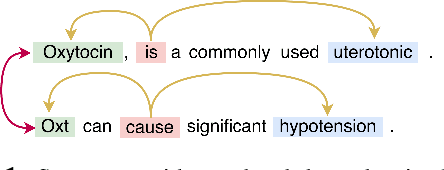
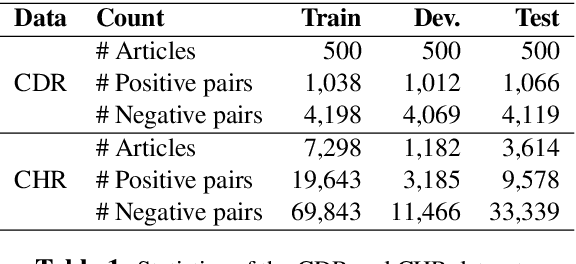
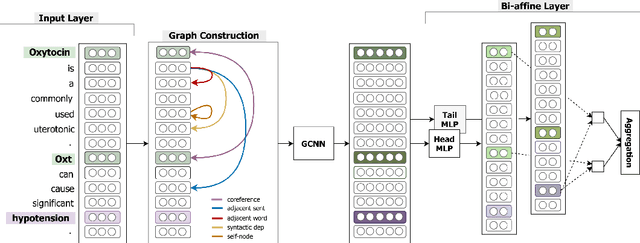

Abstract:Inter-sentence relation extraction deals with a number of complex semantic relationships in documents, which require local, non-local, syntactic and semantic dependencies. Existing methods do not fully exploit such dependencies. We present a novel inter-sentence relation extraction model that builds a labelled edge graph convolutional neural network model on a document-level graph. The graph is constructed using various inter- and intra-sentence dependencies to capture local and non-local dependency information. In order to predict the relation of an entity pair, we utilise multi-instance learning with bi-affine pairwise scoring. Experimental results show that our model achieves comparable performance to the state-of-the-art neural models on two biochemistry datasets. Our analysis shows that all the types in the graph are effective for inter-sentence relation extraction.
Siamese Neural Networks with Random Forest for detecting duplicate question pairs
Jan 28, 2018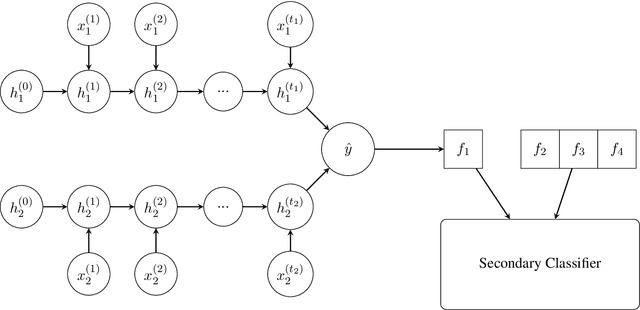



Abstract:Determining whether two given questions are semantically similar is a fairly challenging task given the different structures and forms that the questions can take. In this paper, we use Gated Recurrent Units(GRU) in combination with other highly used machine learning algorithms like Random Forest, Adaboost and SVM for the similarity prediction task on a dataset released by Quora, consisting of about 400k labeled question pairs. We got the best result by using the Siamese adaptation of a Bidirectional GRU with a Random Forest classifier, which landed us among the top 24% in the competition Quora Question Pairs hosted on Kaggle.
What matters in a transferable neural network model for relation classification in the biomedical domain?
Aug 14, 2017



Abstract:Lack of sufficient labeled data often limits the applicability of advanced machine learning algorithms to real life problems. However efficient use of Transfer Learning (TL) has been shown to be very useful across domains. TL utilizes valuable knowledge learned in one task (source task), where sufficient data is available, to the task of interest (target task). In biomedical and clinical domain, it is quite common that lack of sufficient training data do not allow to fully exploit machine learning models. In this work, we present two unified recurrent neural models leading to three transfer learning frameworks for relation classification tasks. We systematically investigate effectiveness of the proposed frameworks in transferring the knowledge under multiple aspects related to source and target tasks, such as, similarity or relatedness between source and target tasks, and size of training data for source task. Our empirical results show that the proposed frameworks in general improve the model performance, however these improvements do depend on aspects related to source and target tasks. This dependence then finally determine the choice of a particular TL framework.
Drug-Drug Interaction Extraction from Biomedical Text Using Long Short Term Memory Network
Aug 13, 2017



Abstract:Simultaneous administration of multiple drugs can have synergistic or antagonistic effects as one drug can affect activities of other drugs. Synergistic effects lead to improved therapeutic outcomes, whereas, antagonistic effects can be life-threatening, may lead to increased healthcare cost, or may even cause death. Thus identification of unknown drug-drug interaction (DDI) is an important concern for efficient and effective healthcare. Although multiple resources for DDI exist, they are often unable to keep pace with rich amount of information available in fast growing biomedical texts. Most existing methods model DDI extraction from text as a classification problem and mainly rely on handcrafted features. Some of these features further depend on domain specific tools. Recently neural network models using latent features have been shown to give similar or better performance than the other existing models dependent on handcrafted features. In this paper, we present three models namely, {\it B-LSTM}, {\it AB-LSTM} and {\it Joint AB-LSTM} based on long short-term memory (LSTM) network. All three models utilize word and position embedding as latent features and thus do not rely on explicit feature engineering. Further use of bidirectional long short-term memory (Bi-LSTM) networks allow implicit feature extraction from the whole sentence. The two models, {\it AB-LSTM} and {\it Joint AB-LSTM} also use attentive pooling in the output of Bi-LSTM layer to assign weights to features. Our experimental results on the SemEval-2013 DDI extraction dataset show that the {\it Joint AB-LSTM} model outperforms all the existing methods, including those relying on handcrafted features. The other two proposed LSTM models also perform competitively with state-of-the-art methods.
Unified Neural Architecture for Drug, Disease and Clinical Entity Recognition
Aug 11, 2017



Abstract:Most existing methods for biomedical entity recognition task rely on explicit feature engineering where many features either are specific to a particular task or depends on output of other existing NLP tools. Neural architectures have been shown across various domains that efforts for explicit feature design can be reduced. In this work we propose an unified framework using bi-directional long short term memory network (BLSTM) for named entity recognition (NER) tasks in biomedical and clinical domains. Three important characteristics of the framework are as follows - (1) model learns contextual as well as morphological features using two different BLSTM in hierarchy, (2) model uses first order linear conditional random field (CRF) in its output layer in cascade of BLSTM to infer label or tag sequence, (3) model does not use any domain specific features or dictionary, i.e., in another words, same set of features are used in the three NER tasks, namely, disease name recognition (Disease NER), drug name recognition (Drug NER) and clinical entity recognition (Clinical NER). We compare performance of the proposed model with existing state-of-the-art models on the standard benchmark datasets of the three tasks. We show empirically that the proposed framework outperforms all existing models. Further our analysis of CRF layer and word-embedding obtained using character based embedding show their importance.
Biomedical Event Trigger Identification Using Bidirectional Recurrent Neural Network Based Models
May 26, 2017



Abstract:Biomedical events describe complex interactions between various biomedical entities. Event trigger is a word or a phrase which typically signifies the occurrence of an event. Event trigger identification is an important first step in all event extraction methods. However many of the current approaches either rely on complex hand-crafted features or consider features only within a window. In this paper we propose a method that takes the advantage of recurrent neural network (RNN) to extract higher level features present across the sentence. Thus hidden state representation of RNN along with word and entity type embedding as features avoid relying on the complex hand-crafted features generated using various NLP toolkits. Our experiments have shown to achieve state-of-art F1-score on Multi Level Event Extraction (MLEE) corpus. We have also performed category-wise analysis of the result and discussed the importance of various features in trigger identification task.
Recurrent neural network models for disease name recognition using domain invariant features
Jun 30, 2016



Abstract:Hand-crafted features based on linguistic and domain-knowledge play crucial role in determining the performance of disease name recognition systems. Such methods are further limited by the scope of these features or in other words, their ability to cover the contexts or word dependencies within a sentence. In this work, we focus on reducing such dependencies and propose a domain-invariant framework for the disease name recognition task. In particular, we propose various end-to-end recurrent neural network (RNN) models for the tasks of disease name recognition and their classification into four pre-defined categories. We also utilize convolution neural network (CNN) in cascade of RNN to get character-based embedded features and employ it with word-embedded features in our model. We compare our models with the state-of-the-art results for the two tasks on NCBI disease dataset. Our results for the disease mention recognition task indicate that state-of-the-art performance can be obtained without relying on feature engineering. Further the proposed models obtained improved performance on the classification task of disease names.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge