Shenggan Cheng
SRDiffusion: Accelerate Video Diffusion Inference via Sketching-Rendering Cooperation
May 25, 2025Abstract:Leveraging the diffusion transformer (DiT) architecture, models like Sora, CogVideoX and Wan have achieved remarkable progress in text-to-video, image-to-video, and video editing tasks. Despite these advances, diffusion-based video generation remains computationally intensive, especially for high-resolution, long-duration videos. Prior work accelerates its inference by skipping computation, usually at the cost of severe quality degradation. In this paper, we propose SRDiffusion, a novel framework that leverages collaboration between large and small models to reduce inference cost. The large model handles high-noise steps to ensure semantic and motion fidelity (Sketching), while the smaller model refines visual details in low-noise steps (Rendering). Experimental results demonstrate that our method outperforms existing approaches, over 3$\times$ speedup for Wan with nearly no quality loss for VBench, and 2$\times$ speedup for CogVideoX. Our method is introduced as a new direction orthogonal to existing acceleration strategies, offering a practical solution for scalable video generation.
DSP: Dynamic Sequence Parallelism for Multi-Dimensional Transformers
Mar 15, 2024



Abstract:Scaling large models with long sequences across applications like language generation, video generation and multimodal tasks requires efficient sequence parallelism. However, existing sequence parallelism methods all assume a single sequence dimension and fail to adapt to multi-dimensional transformer architectures that perform attention calculations across different dimensions. This paper introduces Dynamic Sequence Parallelism (DSP), a novel approach to enable efficient sequence parallelism for multi-dimensional transformer models. The key idea is to dynamically switch the parallelism dimension according to the current computation stage, leveraging the potential characteristics of multi-dimensional attention. This dynamic dimension switching allows sequence parallelism with minimal communication overhead compared to applying traditional single-dimension parallelism to multi-dimensional models. Experiments show DSP improves end-to-end throughput by 42.0% to 216.8% over prior sequence parallelism methods.
AutoChunk: Automated Activation Chunk for Memory-Efficient Long Sequence Inference
Jan 19, 2024



Abstract:Large deep learning models have achieved impressive performance across a range of applications. However, their large memory requirements, including parameter memory and activation memory, have become a significant challenge for their practical serving. While existing methods mainly address parameter memory, the importance of activation memory has been overlooked. Especially for long input sequences, activation memory is expected to experience a significant exponential growth as the length of sequences increases. In this approach, we propose AutoChunk, an automatic and adaptive compiler system that efficiently reduces activation memory for long sequence inference by chunk strategies. The proposed system generates chunk plans by optimizing through multiple stages. In each stage, the chunk search pass explores all possible chunk candidates and the chunk selection pass identifies the optimal one. At runtime, AutoChunk employs code generation to automatically apply chunk strategies. The experiments demonstrate that AutoChunk can reduce over 80\% of activation memory while maintaining speed loss within 10%, extend max sequence length by 3.2x to 11.7x, and outperform state-of-the-art methods by a large margin.
FastFold: Reducing AlphaFold Training Time from 11 Days to 67 Hours
Mar 04, 2022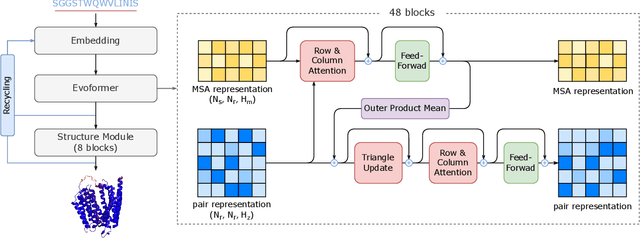
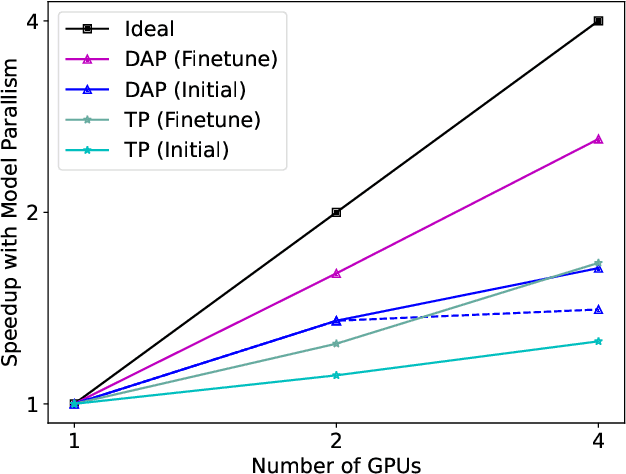
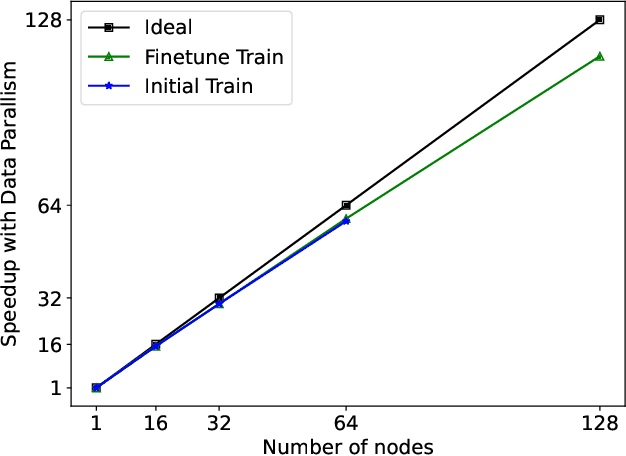
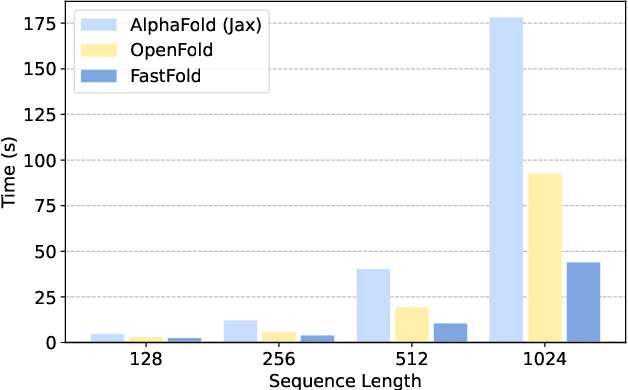
Abstract:Protein structure prediction is an important method for understanding gene translation and protein function in the domain of structural biology. AlphaFold introduced the Transformer model to the field of protein structure prediction with atomic accuracy. However, training and inference of the AlphaFold model are time-consuming and expensive because of the special performance characteristics and huge memory consumption. In this paper, we propose FastFold, a highly efficient implementation of the protein structure prediction model for training and inference. FastFold includes a series of GPU optimizations based on a thorough analysis of AlphaFold's performance. Meanwhile, with Dynamic Axial Parallelism and Duality Async Operation, FastFold achieves high model parallelism scaling efficiency, surpassing existing popular model parallelism techniques. Experimental results show that FastFold reduces overall training time from 11 days to 67 hours and achieves 7.5-9.5X speedup for long-sequence inference. Furthermore, We scaled FastFold to 512 GPUs and achieved an aggregate of 6.02 PetaFLOPs with 90.1% parallel efficiency. The implementation can be found at https://github.com/hpcaitech/FastFold
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge