Ruidong Wu
FAFE: Immune Complex Modeling with Geodesic Distance Loss on Noisy Group Frames
Jul 01, 2024



Abstract:Despite the striking success of general protein folding models such as AlphaFold2(AF2, Jumper et al. (2021)), the accurate computational modeling of antibody-antigen complexes remains a challenging task. In this paper, we first analyze AF2's primary loss function, known as the Frame Aligned Point Error (FAPE), and raise a previously overlooked issue that FAPE tends to face gradient vanishing problem on high-rotational-error targets. To address this fundamental limitation, we propose a novel geodesic loss called Frame Aligned Frame Error (FAFE, denoted as F2E to distinguish from FAPE), which enables the model to better optimize both the rotational and translational errors between two frames. We then prove that F2E can be reformulated as a group-aware geodesic loss, which translates the optimization of the residue-to-residue error to optimizing group-to-group geodesic frame distance. By fine-tuning AF2 with our proposed new loss function, we attain a correct rate of 52.3\% (DockQ $>$ 0.23) on an evaluation set and 43.8\% correct rate on a subset with low homology, with substantial improvement over AF2 by 182\% and 100\% respectively.
FastFold: Reducing AlphaFold Training Time from 11 Days to 67 Hours
Mar 04, 2022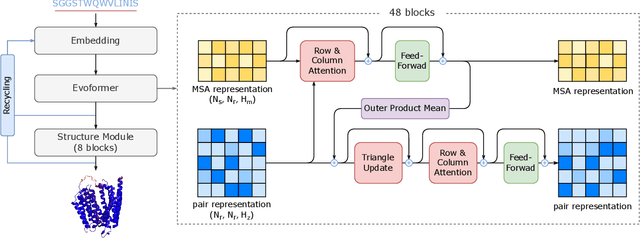
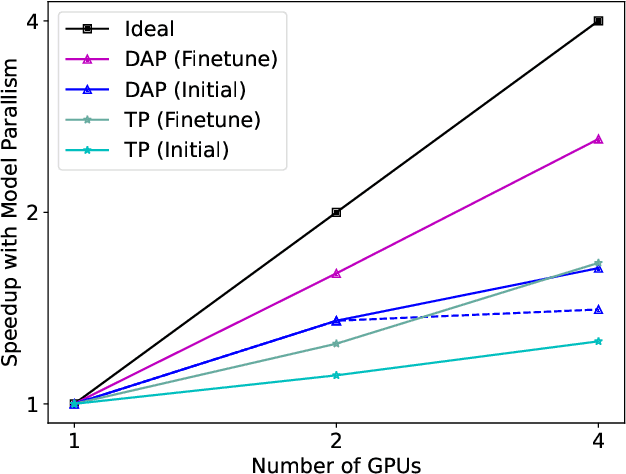
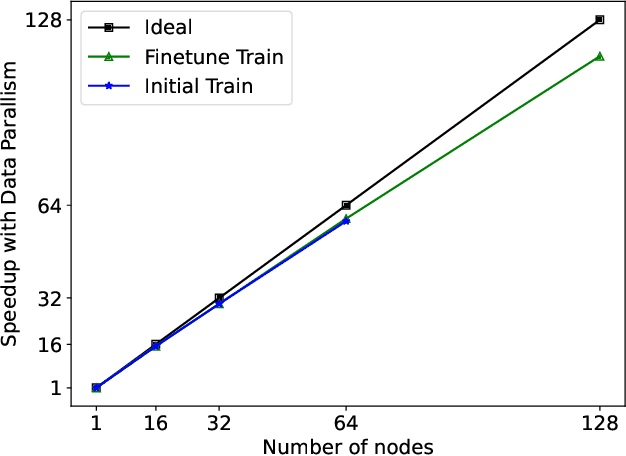
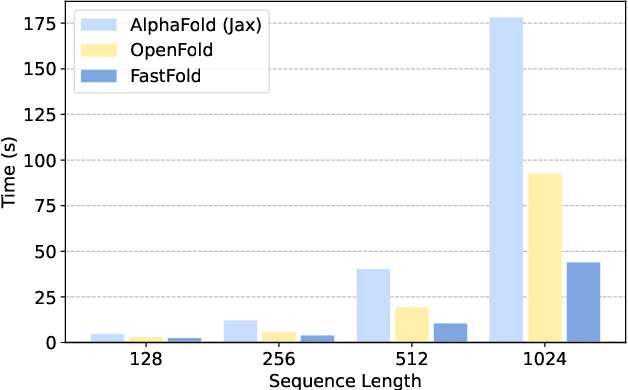
Abstract:Protein structure prediction is an important method for understanding gene translation and protein function in the domain of structural biology. AlphaFold introduced the Transformer model to the field of protein structure prediction with atomic accuracy. However, training and inference of the AlphaFold model are time-consuming and expensive because of the special performance characteristics and huge memory consumption. In this paper, we propose FastFold, a highly efficient implementation of the protein structure prediction model for training and inference. FastFold includes a series of GPU optimizations based on a thorough analysis of AlphaFold's performance. Meanwhile, with Dynamic Axial Parallelism and Duality Async Operation, FastFold achieves high model parallelism scaling efficiency, surpassing existing popular model parallelism techniques. Experimental results show that FastFold reduces overall training time from 11 days to 67 hours and achieves 7.5-9.5X speedup for long-sequence inference. Furthermore, We scaled FastFold to 512 GPUs and achieved an aggregate of 6.02 PetaFLOPs with 90.1% parallel efficiency. The implementation can be found at https://github.com/hpcaitech/FastFold
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge