Shahin Boluki
Machine Learning based Framework for Robust Price-Sensitivity Estimation with Application to Airline Pricing
May 04, 2022



Abstract:We consider the problem of dynamic pricing of a product in the presence of feature-dependent price sensitivity. Based on the Poisson semi-parametric approach, we construct a flexible yet interpretable demand model where the price related part is parametric while the remaining (nuisance) part of the model is non-parametric and can be modeled via sophisticated ML techniques. The estimation of price-sensitivity parameters of this model via direct one-stage regression techniques may lead to biased estimates. We propose a two-stage estimation methodology which makes the estimation of the price-sensitivity parameters robust to biases in the nuisance parameters of the model. In the first-stage we construct the estimators of observed purchases and price given the feature vector using sophisticated ML estimators like deep neural networks. Utilizing the estimators from the first-stage, in the second-stage we leverage a Bayesian dynamic generalized linear model to estimate the price-sensitivity parameters. We test the performance of the proposed estimation schemes on simulated and real sales transaction data from Airline industry. Our numerical studies demonstrate that the two-stage approach provides more accurate estimates of price-sensitivity parameters as compared to direct one-stage approach.
VFDS: Variational Foresight Dynamic Selection in Bayesian Neural Networks for Efficient Human Activity Recognition
Mar 31, 2022



Abstract:In many machine learning tasks, input features with varying degrees of predictive capability are acquired at varying costs. In order to optimize the performance-cost trade-off, one would select features to observe a priori. However, given the changing context with previous observations, the subset of predictive features to select may change dynamically. Therefore, we face the challenging new problem of foresight dynamic selection (FDS): finding a dynamic and light-weight policy to decide which features to observe next, before actually observing them, for overall performance-cost trade-offs. To tackle FDS, this paper proposes a Bayesian learning framework of Variational Foresight Dynamic Selection (VFDS). VFDS learns a policy that selects the next feature subset to observe, by optimizing a variational Bayesian objective that characterizes the trade-off between model performance and feature cost. At its core is an implicit variational distribution on binary gates that are dependent on previous observations, which will select the next subset of features to observe. We apply VFDS on the Human Activity Recognition (HAR) task where the performance-cost trade-off is critical in its practice. Extensive results demonstrate that VFDS selects different features under changing contexts, notably saving sensory costs while maintaining or improving the HAR accuracy. Moreover, the features that VFDS dynamically select are shown to be interpretable and associated with the different activity types. We will release the code.
SimCD: Simultaneous Clustering and Differential expression analysis for single-cell transcriptomic data
Apr 04, 2021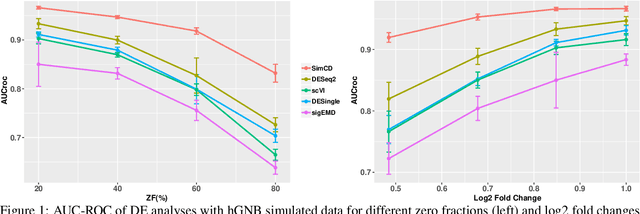
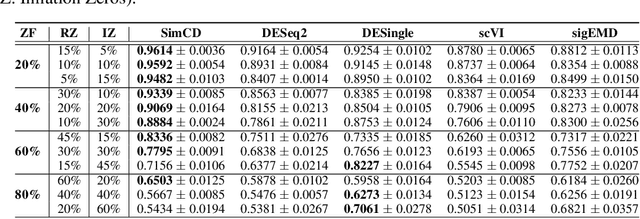
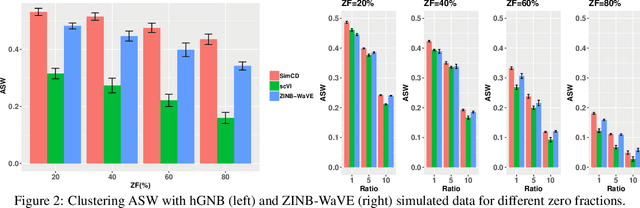
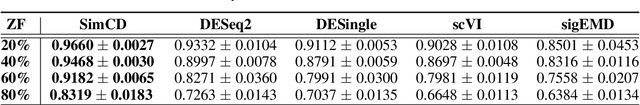
Abstract:Single-Cell RNA sequencing (scRNA-seq) measurements have facilitated genome-scale transcriptomic profiling of individual cells, with the hope of deconvolving cellular dynamic changes in corresponding cell sub-populations to better understand molecular mechanisms of different development processes. Several scRNA-seq analysis methods have been proposed to first identify cell sub-populations by clustering and then separately perform differential expression analysis to understand gene expression changes. Their corresponding statistical models and inference algorithms are often designed disjointly. We develop a new method -- SimCD -- that explicitly models cell heterogeneity and dynamic differential changes in one unified hierarchical gamma-negative binomial (hGNB) model, allowing simultaneous cell clustering and differential expression analysis for scRNA-seq data. Our method naturally defines cell heterogeneity by dynamic expression changes, which is expected to help achieve better performances on the two tasks compared to the existing methods that perform them separately. In addition, SimCD better models dropout (zero inflation) in scRNA-seq data by both cell- and gene-level factors and obviates the need for sophisticated pre-processing steps such as normalization, thanks to the direct modeling of scRNA-seq count data by the rigorous hGNB model with an efficient Gibbs sampling inference algorithm. Extensive comparisons with the state-of-the-art methods on both simulated and real-world scRNA-seq count data demonstrate the capability of SimCD to discover cell clusters and capture dynamic expression changes. Furthermore, SimCD helps identify several known genes affected by food deprivation in hypothalamic neuron cell subtypes as well as some new potential markers, suggesting the capability of SimCD for bio-marker discovery.
Bayesian Graph Neural Networks with Adaptive Connection Sampling
Jun 30, 2020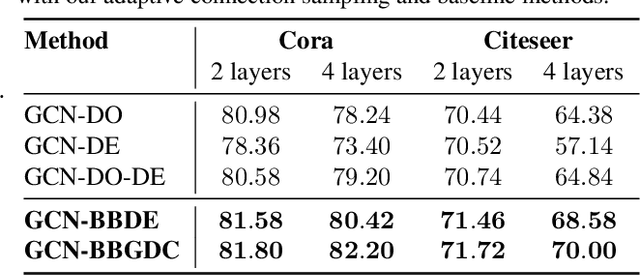
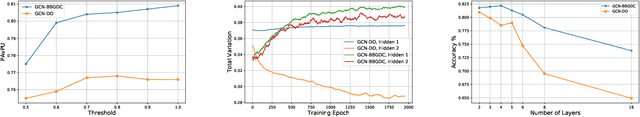
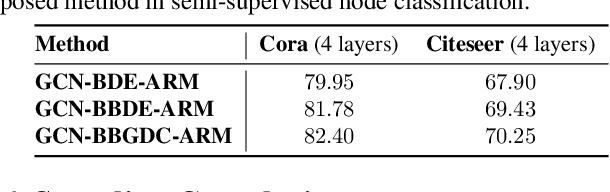
Abstract:We propose a unified framework for adaptive connection sampling in graph neural networks (GNNs) that generalizes existing stochastic regularization methods for training GNNs. The proposed framework not only alleviates over-smoothing and over-fitting tendencies of deep GNNs, but also enables learning with uncertainty in graph analytic tasks with GNNs. Instead of using fixed sampling rates or hand-tuning them as model hyperparameters in existing stochastic regularization methods, our adaptive connection sampling can be trained jointly with GNN model parameters in both global and local fashions. GNN training with adaptive connection sampling is shown to be mathematically equivalent to an efficient approximation of training Bayesian GNNs. Experimental results with ablation studies on benchmark datasets validate that adaptively learning the sampling rate given graph training data is the key to boost the performance of GNNs in semi-supervised node classification, less prone to over-smoothing and over-fitting with more robust prediction.
NADS: Neural Architecture Distribution Search for Uncertainty Awareness
Jun 11, 2020



Abstract:Machine learning (ML) systems often encounter Out-of-Distribution (OoD) errors when dealing with testing data coming from a distribution different from training data. It becomes important for ML systems in critical applications to accurately quantify its predictive uncertainty and screen out these anomalous inputs. However, existing OoD detection approaches are prone to errors and even sometimes assign higher likelihoods to OoD samples. Unlike standard learning tasks, there is currently no well established guiding principle for designing OoD detection architectures that can accurately quantify uncertainty. To address these problems, we first seek to identify guiding principles for designing uncertainty-aware architectures, by proposing Neural Architecture Distribution Search (NADS). NADS searches for a distribution of architectures that perform well on a given task, allowing us to identify common building blocks among all uncertainty-aware architectures. With this formulation, we are able to optimize a stochastic OoD detection objective and construct an ensemble of models to perform OoD detection. We perform multiple OoD detection experiments and observe that our NADS performs favorably, with up to 57% improvement in accuracy compared to state-of-the-art methods among 15 different testing configurations.
Pairwise Supervised Hashing with Bernoulli Variational Auto-Encoder and Self-Control Gradient Estimator
May 21, 2020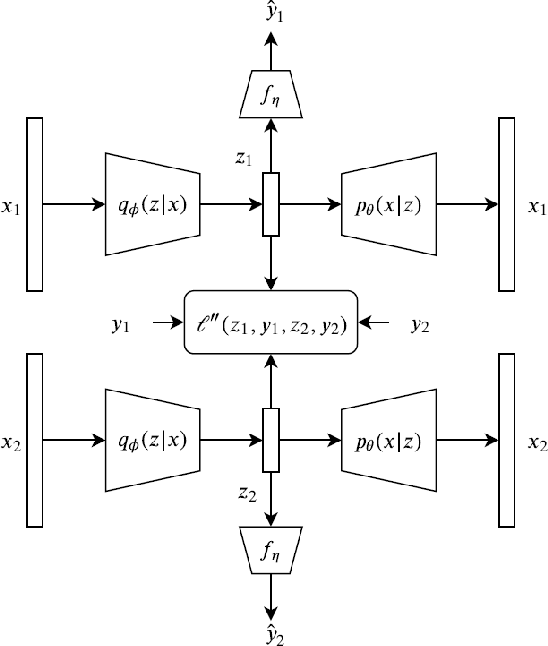
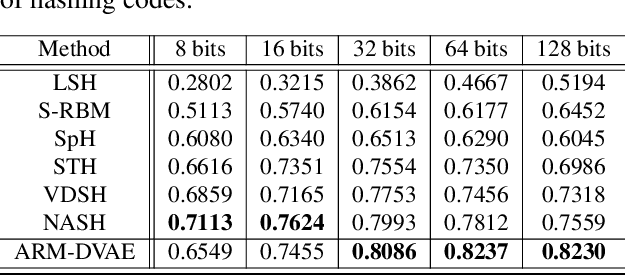
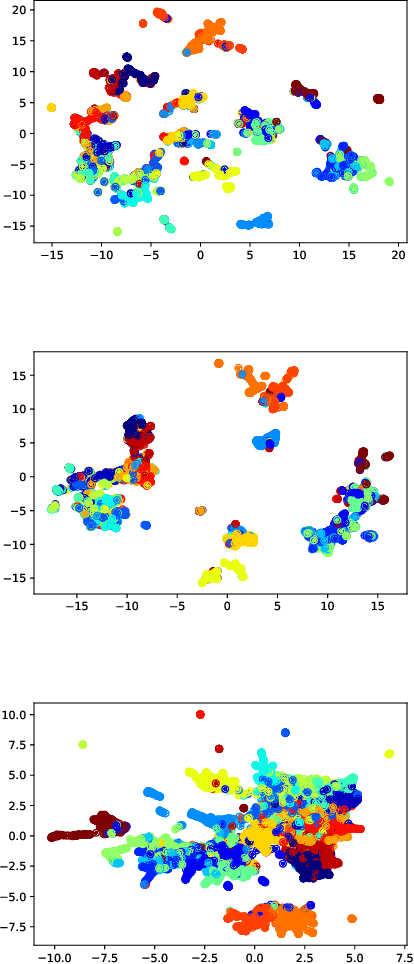
Abstract:Semantic hashing has become a crucial component of fast similarity search in many large-scale information retrieval systems, in particular, for text data. Variational auto-encoders (VAEs) with binary latent variables as hashing codes provide state-of-the-art performance in terms of precision for document retrieval. We propose a pairwise loss function with discrete latent VAE to reward within-class similarity and between-class dissimilarity for supervised hashing. Instead of solving the optimization relying on existing biased gradient estimators, an unbiased low-variance gradient estimator is adopted to optimize the hashing function by evaluating the non-differentiable loss function over two correlated sets of binary hashing codes to control the variance of gradient estimates. This new semantic hashing framework achieves superior performance compared to the state-of-the-arts, as demonstrated by our comprehensive experiments.
* To appear in UAI 2020
Learnable Bernoulli Dropout for Bayesian Deep Learning
Feb 12, 2020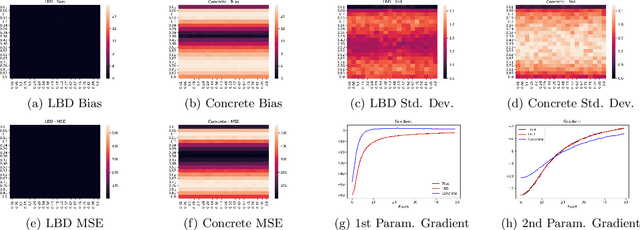
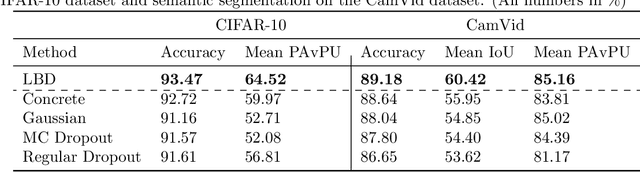
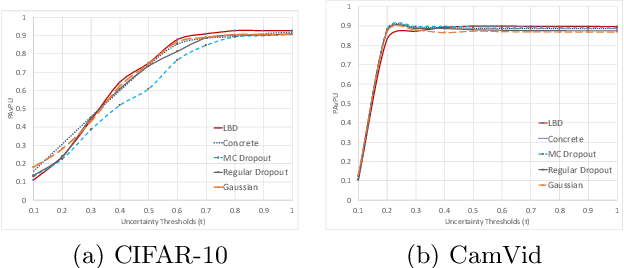
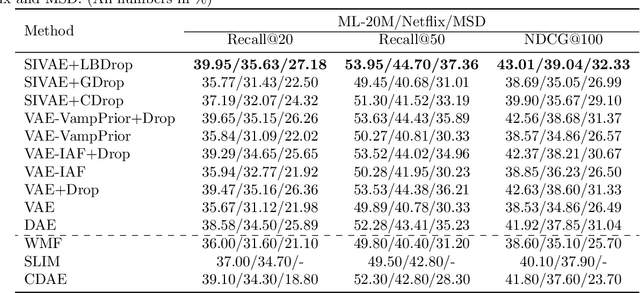
Abstract:In this work, we propose learnable Bernoulli dropout (LBD), a new model-agnostic dropout scheme that considers the dropout rates as parameters jointly optimized with other model parameters. By probabilistic modeling of Bernoulli dropout, our method enables more robust prediction and uncertainty quantification in deep models. Especially, when combined with variational auto-encoders (VAEs), LBD enables flexible semi-implicit posterior representations, leading to new semi-implicit VAE~(SIVAE) models. We solve the optimization for training with respect to the dropout parameters using Augment-REINFORCE-Merge (ARM), an unbiased and low-variance gradient estimator. Our experiments on a range of tasks show the superior performance of our approach compared with other commonly used dropout schemes. Overall, LBD leads to improved accuracy and uncertainty estimates in image classification and semantic segmentation. Moreover, using SIVAE, we can achieve state-of-the-art performance on collaborative filtering for implicit feedback on several public datasets.
ARSM Gradient Estimator for Supervised Learning to Rank
Nov 01, 2019
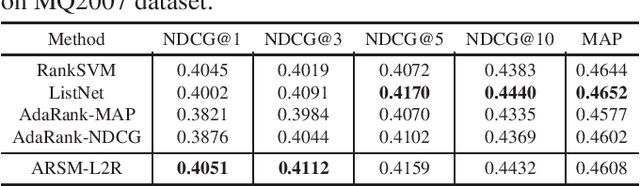
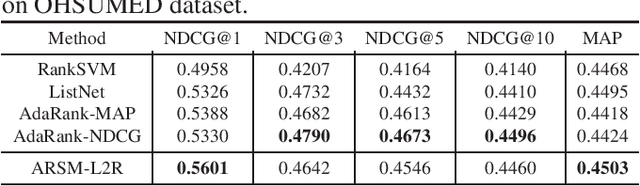
Abstract:We propose a new model for supervised learning to rank. In our model, the relevancy labels are are assumed to follow a categorical distribution whose probabilities are constructed based on a scoring function. We optimize the training objective with respect to the multivariate categorical variables with an unbiased and low-variance gradient estimator. Learning to rank methods can generally be categorized into pointwise, pairwise, and listwise approaches. Our approach belongs to the class of pointwise methods. Although it has previously been reported that pointwise methods cannot achieve as good performance as of pairwise or listwise approaches, we show that the proposed method achieves better or comparable results on two datasets compared with pairwise and listwise methods.
Optimal Clustering with Missing Values
Feb 26, 2019
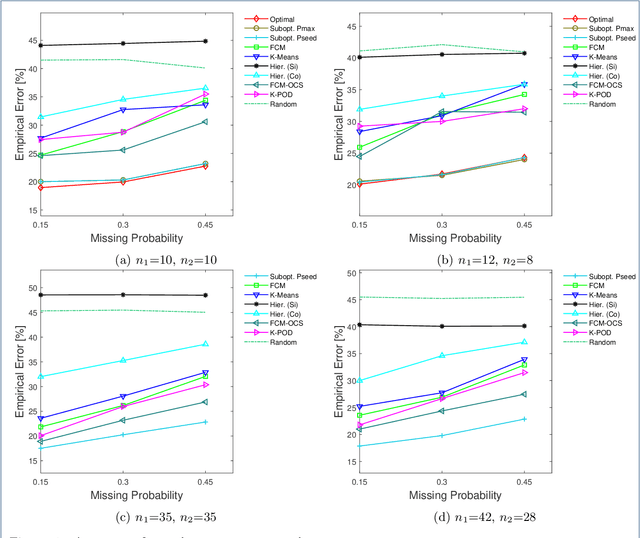
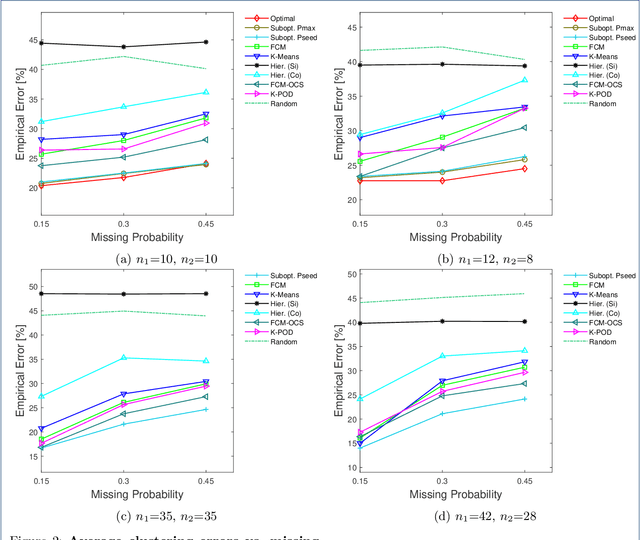
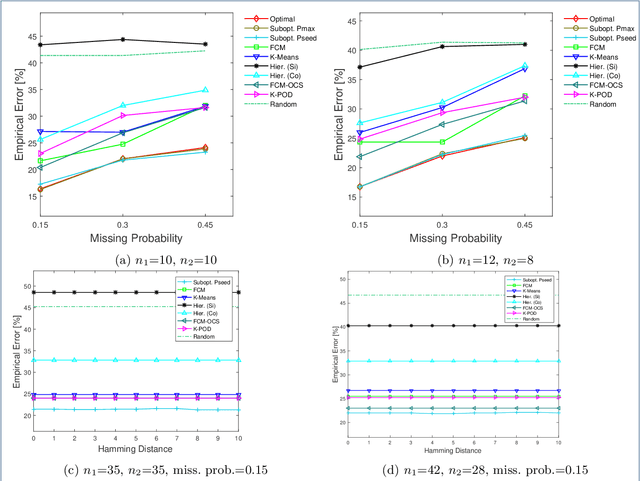
Abstract:Missing values frequently arise in modern biomedical studies due to various reasons, including missing tests or complex profiling technologies for different omics measurements. Missing values can complicate the application of clustering algorithms, whose goals are to group points based on some similarity criterion. A common practice for dealing with missing values in the context of clustering is to first impute the missing values, and then apply the clustering algorithm on the completed data. We consider missing values in the context of optimal clustering, which finds an optimal clustering operator with reference to an underlying random labeled point process (RLPP). We show how the missing-value problem fits neatly into the overall framework of optimal clustering by incorporating the missing value mechanism into the random labeled point process and then marginalizing out the missing-value process. In particular, we demonstrate the proposed framework for the Gaussian model with arbitrary covariance structures. Comprehensive experimental studies on both synthetic and real-world RNA-seq data show the superior performance of the proposed optimal clustering with missing values when compared to various clustering approaches. Optimal clustering with missing values obviates the need for imputation-based pre-processing of the data, while at the same time possessing smaller clustering errors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge