Semih Cantürk
Towards Graph Foundation Models: A Study on the Generalization of Positional and Structural Encodings
Dec 10, 2024Abstract:Recent advances in integrating positional and structural encodings (PSEs) into graph neural networks (GNNs) have significantly enhanced their performance across various graph learning tasks. However, the general applicability of these encodings and their potential to serve as foundational representations for graphs remain uncertain. This paper investigates the fine-tuning efficiency, scalability with sample size, and generalization capability of learnable PSEs across diverse graph datasets. Specifically, we evaluate their potential as universal pre-trained models that can be easily adapted to new tasks with minimal fine-tuning and limited data. Furthermore, we assess the expressivity of the learned representations, particularly, when used to augment downstream GNNs. We demonstrate through extensive benchmarking and empirical analysis that PSEs generally enhance downstream models. However, some datasets may require specific PSE-augmentations to achieve optimal performance. Nevertheless, our findings highlight their significant potential to become integral components of future graph foundation models. We provide new insights into the strengths and limitations of PSEs, contributing to the broader discourse on foundation models in graph learning.
Towards a General GNN Framework for Combinatorial Optimization
May 31, 2024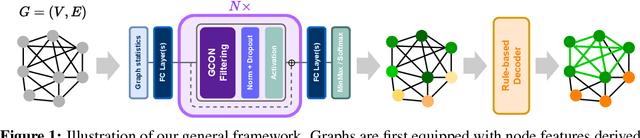
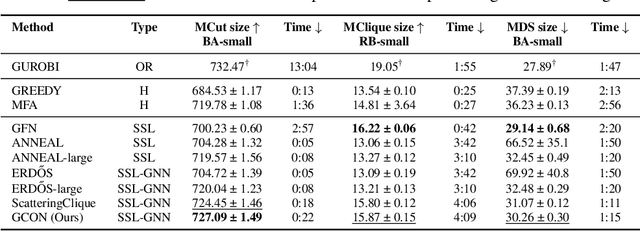
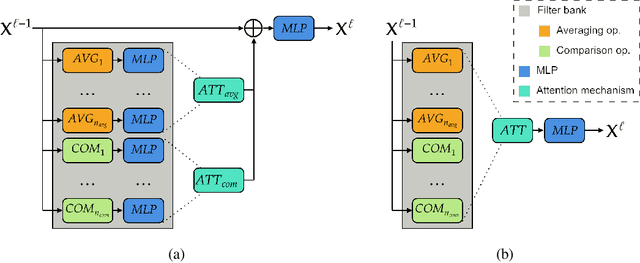
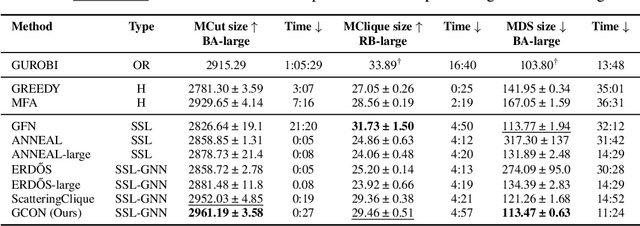
Abstract:Graph neural networks (GNNs) have achieved great success for a variety of tasks such as node classification, graph classification, and link prediction. However, the use of GNNs (and machine learning more generally) to solve combinatorial optimization (CO) problems is much less explored. Here, we introduce a novel GNN architecture which leverages a complex filter bank and localized attention mechanisms designed to solve CO problems on graphs. We show how our method differentiates itself from prior GNN-based CO solvers and how it can be effectively applied to the maximum clique, minimum dominating set, and maximum cut problems in a self-supervised learning setting. In addition to demonstrating competitive overall performance across all tasks, we establish state-of-the-art results for the max cut problem.
Graph Positional and Structural Encoder
Jul 14, 2023



Abstract:Positional and structural encodings (PSE) enable better identifiability of nodes within a graph, as in general graphs lack a canonical node ordering. This renders PSEs essential tools for empowering modern GNNs, and in particular graph Transformers. However, designing PSEs that work optimally for a variety of graph prediction tasks is a challenging and unsolved problem. Here, we present the graph positional and structural encoder (GPSE), a first-ever attempt to train a graph encoder that captures rich PSE representations for augmenting any GNN. GPSE can effectively learn a common latent representation for multiple PSEs, and is highly transferable. The encoder trained on a particular graph dataset can be used effectively on datasets drawn from significantly different distributions and even modalities. We show that across a wide range of benchmarks, GPSE-enhanced models can significantly improve the performance in certain tasks, while performing on par with those that employ explicitly computed PSEs in other cases. Our results pave the way for the development of large pre-trained models for extracting graph positional and structural information and highlight their potential as a viable alternative to explicitly computed PSEs as well as to existing self-supervised pre-training approaches.
Taxonomy of Benchmarks in Graph Representation Learning
Jun 15, 2022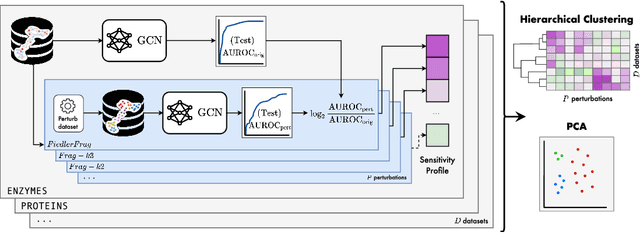
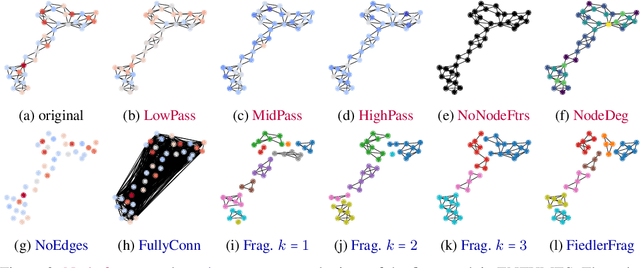
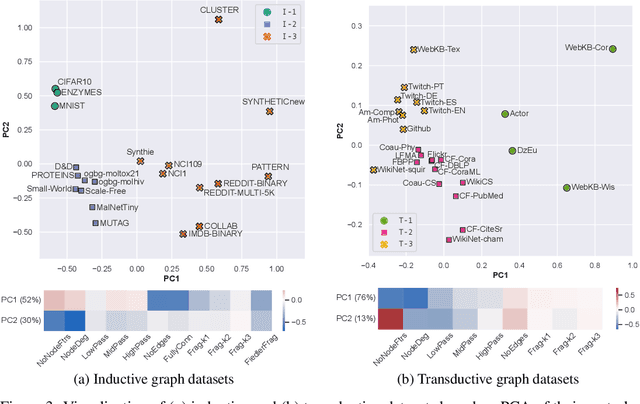
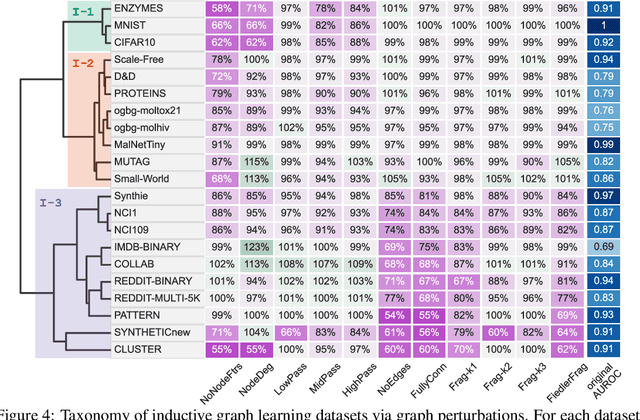
Abstract:Graph Neural Networks (GNNs) extend the success of neural networks to graph-structured data by accounting for their intrinsic geometry. While extensive research has been done on developing GNN models with superior performance according to a collection of graph representation learning benchmarks, it is currently not well understood what aspects of a given model are probed by them. For example, to what extent do they test the ability of a model to leverage graph structure vs. node features? Here, we develop a principled approach to taxonomize benchmarking datasets according to a $\textit{sensitivity profile}$ that is based on how much GNN performance changes due to a collection of graph perturbations. Our data-driven analysis provides a deeper understanding of which benchmarking data characteristics are leveraged by GNNs. Consequently, our taxonomy can aid in selection and development of adequate graph benchmarks, and better informed evaluation of future GNN methods. Finally, our approach and implementation in $\texttt{GTaxoGym}$ package are extendable to multiple graph prediction task types and future datasets.
Towards a Taxonomy of Graph Learning Datasets
Oct 27, 2021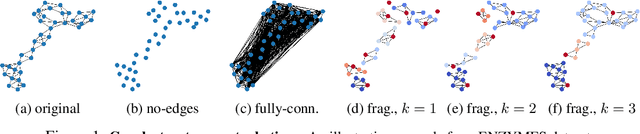
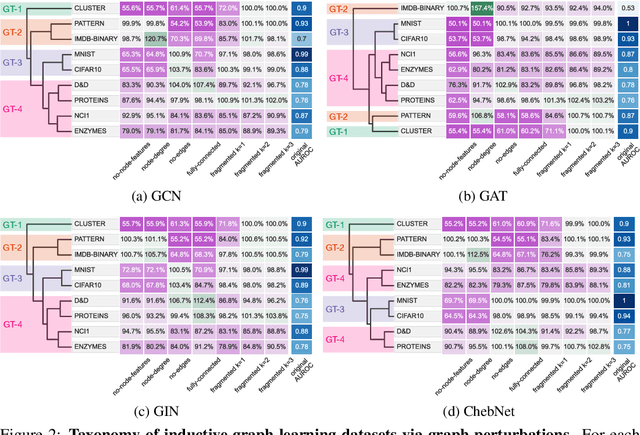
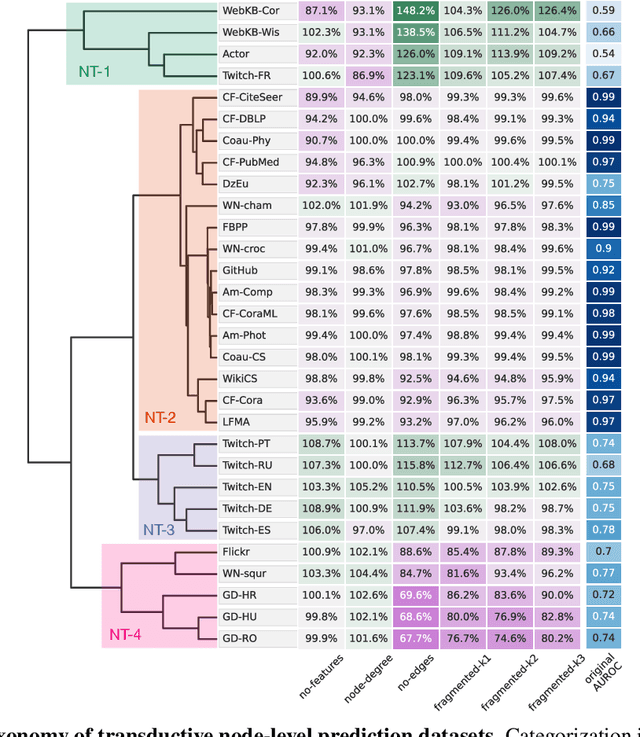
Abstract:Graph neural networks (GNNs) have attracted much attention due to their ability to leverage the intrinsic geometries of the underlying data. Although many different types of GNN models have been developed, with many benchmarking procedures to demonstrate the superiority of one GNN model over the others, there is a lack of systematic understanding of the underlying benchmarking datasets, and what aspects of the model are being tested. Here, we provide a principled approach to taxonomize graph benchmarking datasets by carefully designing a collection of graph perturbations to probe the essential data characteristics that GNN models leverage to perform predictions. Our data-driven taxonomization of graph datasets provides a new understanding of critical dataset characteristics that will enable better model evaluation and the development of more specialized GNN models.
Machine-Learning Driven Drug Repurposing for COVID-19
Jun 25, 2020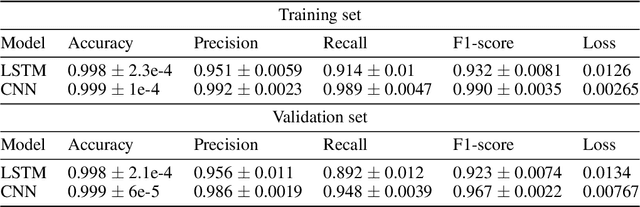
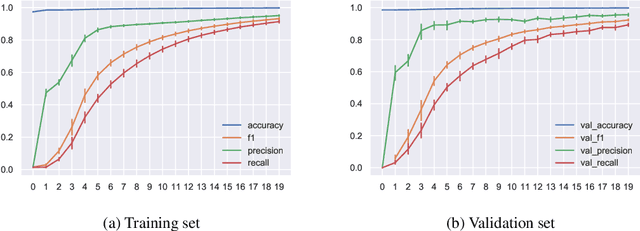
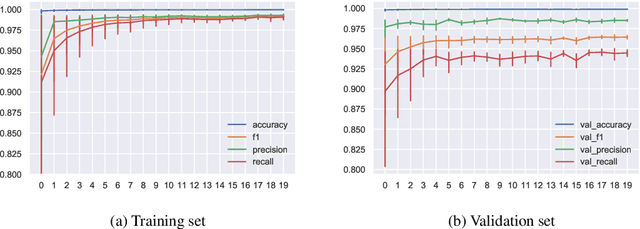
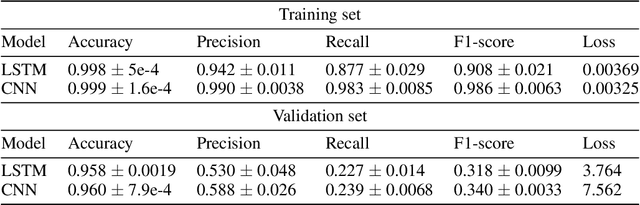
Abstract:The integration of machine learning methods into bioinformatics provides particular benefits in identifying how therapeutics effective in one context might have utility in an unknown clinical context or against a novel pathology. We aim to discover the underlying associations between viral proteins and antiviral therapeutics that are effective against them by employing neural network models. Using the National Center for Biotechnology Information virus protein database and the DrugVirus database, which provides a comprehensive report of broad-spectrum antiviral agents (BSAAs) and viruses they inhibit, we trained ANN models with virus protein sequences as inputs and antiviral agents deemed safe-in-humans as outputs. Model training excluded SARS-CoV-2 proteins and included only Phases II, III, IV and Approved level drugs. Using sequences for SARS-CoV-2 (the coronavirus that causes COVID-19) as inputs to the trained models produces outputs of tentative safe-in-human antiviral candidates for treating COVID-19. Our results suggest multiple drug candidates, some of which complement recent findings from noteworthy clinical studies. Our in-silico approach to drug repurposing has promise in identifying new drug candidates and treatments for other viruses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge