Aman Singh
COMPAct: Computational Optimization and Automated Modular design of Planetary Actuators
Oct 08, 2025



Abstract:The optimal design of robotic actuators is a critical area of research, yet limited attention has been given to optimizing gearbox parameters and automating actuator CAD. This paper introduces COMPAct: Computational Optimization and Automated Modular Design of Planetary Actuators, a framework that systematically identifies optimal gearbox parameters for a given motor across four gearbox types, single-stage planetary gearbox (SSPG), compound planetary gearbox (CPG), Wolfrom planetary gearbox (WPG), and double-stage planetary gearbox (DSPG). The framework minimizes mass and actuator width while maximizing efficiency, and further automates actuator CAD generation to enable direct 3D printing without manual redesign. Using this framework, optimal gearbox designs are explored over a wide range of gear ratios, providing insights into the suitability of different gearbox types across various gear ratio ranges. In addition, the framework is used to generate CAD models of all four gearbox types with varying gear ratios and motors. Two actuator types are fabricated and experimentally evaluated through power efficiency, no-load backlash, and transmission stiffness tests. Experimental results indicate that the SSPG actuator achieves a mechanical efficiency of 60-80 %, a no-load backlash of 0.59 deg, and a transmission stiffness of 242.7 Nm/rad, while the CPG actuator demonstrates 60 % efficiency, 2.6 deg backlash, and a stiffness of 201.6 Nm/rad. Code available at: https://anonymous.4open.science/r/COMPAct-SubNum-3408 Video: https://youtu.be/99zOKgxsDho
Spiritual-LLM : Gita Inspired Mental Health Therapy In the Era of LLMs
Jun 23, 2025Abstract:Traditional mental health support systems often generate responses based solely on the user's current emotion and situations, resulting in superficial interventions that fail to address deeper emotional needs. This study introduces a novel framework by integrating spiritual wisdom from the Bhagavad Gita with advanced large language model GPT-4o to enhance emotional well-being. We present the GITes (Gita Integrated Therapy for Emotional Support) dataset, which enhances the existing ExTES mental health dataset by including 10,729 spiritually guided responses generated by GPT-4o and evaluated by domain experts. We benchmark GITes against 12 state-of-the-art LLMs, including both mental health specific and general purpose models. To evaluate spiritual relevance in generated responses beyond what conventional n-gram based metrics capture, we propose a novel Spiritual Insight metric and automate assessment via an LLM as jury framework using chain-of-thought prompting. Integrating spiritual guidance into AI driven support enhances both NLP and spiritual metrics for the best performing LLM Phi3-Mini 3.2B Instruct, achieving improvements of 122.71% in ROUGE, 126.53% in METEOR, 8.15% in BERT score, 15.92% in Spiritual Insight, 18.61% in Sufficiency and 13.22% in Relevance compared to its zero-shot counterpart. While these results reflect substantial improvements across automated empathy and spirituality metrics, further validation in real world patient populations remains a necessary step. Our findings indicate a strong potential for AI systems enriched with spiritual guidance to enhance user satisfaction and perceived support outcomes. The code and dataset will be publicly available to advance further research in this emerging area.
A Chain-Driven, Sandwich-Legged Quadruped Robot: Design and Experimental Analysis
Mar 18, 2025



Abstract:This paper introduces a chain-driven, sandwich-legged, mid-size quadruped robot designed as an accessible research platform. The design prioritizes enhanced locomotion capabilities, improved reliability and safety of the actuation system, and simplified, cost-effective manufacturing processes. Locomotion performance is optimized through a sandwiched leg design and a dual-motor configuration, reducing leg inertia for agile movements. Reliability and safety are achieved by integrating robust cable strain reliefs, efficient heat sinks for motor thermal management, and mechanical limits to restrict leg motion. Simplified design considerations include a quasi-direct drive (QDD) actuator and the adoption of low-cost fabrication techniques, such as laser cutting and 3D printing, to minimize cost and ensure rapid prototyping. The robot weighs approximately 25 kg and is developed at a cost under \$8000, making it a scalable and affordable solution for robotics research. Experimental validations demonstrate the platform's capability to execute trot and crawl gaits on flat terrain and slopes, highlighting its potential as a versatile and reliable quadruped research platform.
Reducing Onboard Processing Time for Path Planning in Dynamically Evolving Polygonal Maps
May 08, 2023



Abstract:Autonomous agents face the challenge of coordinating multiple tasks (perception, motion planning, controller) which are computationally expensive on a single onboard computer. To utilize the onboard processing capacity optimally, it is imperative to arrive at computationally efficient algorithms for global path planning. In this work, it is attempted to reduce the processing time for global path planning in dynamically evolving polygonal maps. In dynamic environments, maps may not remain valid for long. Hence it is of utmost importance to obtain the shortest path quickly in an ever-changing environment. To address this, an existing rapid path-finding algorithm, the Minimal Construct was used. This algorithm discovers only a necessary portion of the Visibility Graph around obstacles and computes collision tests only for lines that seem heuristically promising. Simulations show that this algorithm finds shortest paths faster than traditional grid-based A* searches in most cases, resulting in smoother and shorter paths even in dynamic environments.
Machine-Learning Driven Drug Repurposing for COVID-19
Jun 25, 2020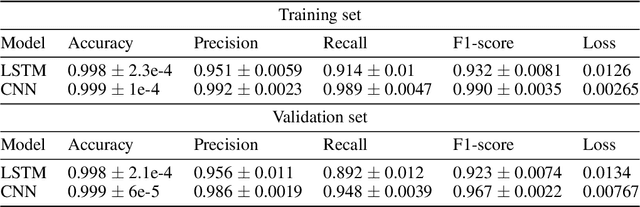
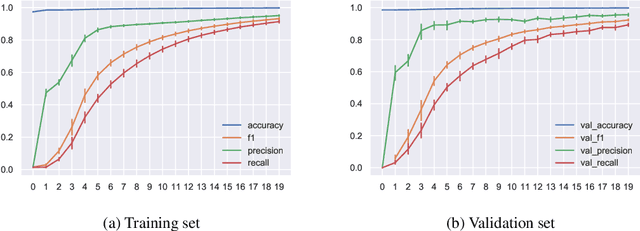
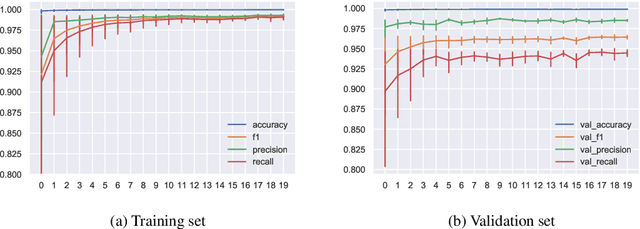
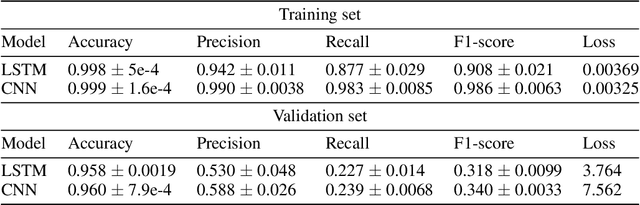
Abstract:The integration of machine learning methods into bioinformatics provides particular benefits in identifying how therapeutics effective in one context might have utility in an unknown clinical context or against a novel pathology. We aim to discover the underlying associations between viral proteins and antiviral therapeutics that are effective against them by employing neural network models. Using the National Center for Biotechnology Information virus protein database and the DrugVirus database, which provides a comprehensive report of broad-spectrum antiviral agents (BSAAs) and viruses they inhibit, we trained ANN models with virus protein sequences as inputs and antiviral agents deemed safe-in-humans as outputs. Model training excluded SARS-CoV-2 proteins and included only Phases II, III, IV and Approved level drugs. Using sequences for SARS-CoV-2 (the coronavirus that causes COVID-19) as inputs to the trained models produces outputs of tentative safe-in-human antiviral candidates for treating COVID-19. Our results suggest multiple drug candidates, some of which complement recent findings from noteworthy clinical studies. Our in-silico approach to drug repurposing has promise in identifying new drug candidates and treatments for other viruses.
SynGAN: Towards Generating Synthetic Network Attacks using GANs
Aug 26, 2019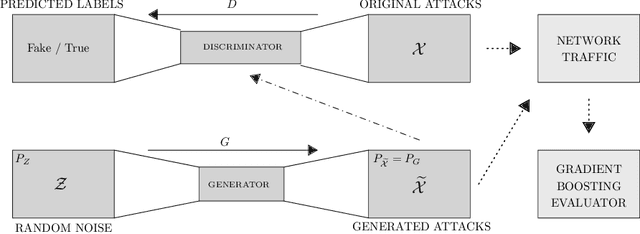

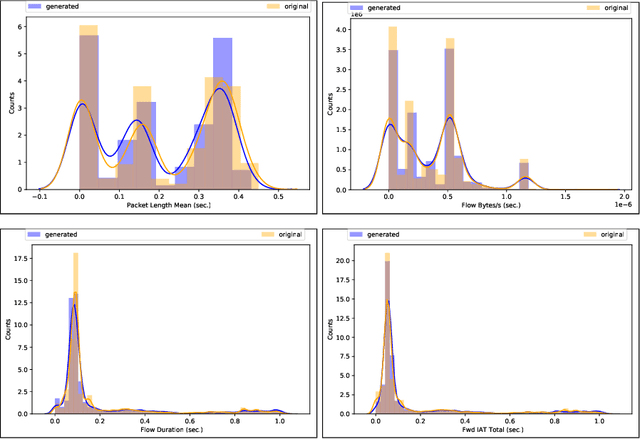
Abstract:The rapid digital transformation without security considerations has resulted in the rise of global-scale cyberattacks. The first line of defense against these attacks are Network Intrusion Detection Systems (NIDS). Once deployed, however, these systems work as blackboxes with a high rate of false positives with no measurable effectiveness. There is a need to continuously test and improve these systems by emulating real-world network attack mutations. We present SynGAN, a framework that generates adversarial network attacks using the Generative Adversial Networks (GAN). SynGAN generates malicious packet flow mutations using real attack traffic, which can improve NIDS attack detection rates. As a first step, we compare two public datasets, NSL-KDD and CICIDS2017, for generating synthetic Distributed Denial of Service (DDoS) network attacks. We evaluate the attack quality (real vs. synthetic) using a gradient boosting classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge