Patrick St-Amant
Clarifying Myths About the Relationship Between Shape Bias, Accuracy, and Robustness
Jun 07, 2024



Abstract:Deep learning models can perform well when evaluated on images from the same distribution as the training set. However, applying small perturbations in the forms of noise, artifacts, occlusions, blurring, etc. to a model's input image and feeding the model with out-of-distribution (OOD) data can significantly drop the model's accuracy, making it not applicable to real-world scenarios. Data augmentation is one of the well-practiced methods to improve model robustness against OOD data; however, examining which augmentation type to choose and how it affects the OOD robustness remains understudied. There is a growing belief that augmenting datasets using data augmentations that improve a model's bias to shape-based features rather than texture-based features results in increased OOD robustness for Convolutional Neural Networks trained on the ImageNet-1K dataset. This is usually stated as ``an increase in the model's shape bias results in an increase in its OOD robustness". Based on this hypothesis, some works in the literature aim to find augmentations with higher effects on model shape bias and use those for data augmentation. By evaluating 39 types of data augmentations on a widely used OOD dataset, we demonstrate the impact of each data augmentation on the model's robustness to OOD data and further show that the mentioned hypothesis is not true; an increase in shape bias does not necessarily result in higher OOD robustness. By analyzing the results, we also find some biases in the ImageNet-1K dataset that can easily be reduced using proper data augmentation. Our evaluation results further show that there is not necessarily a trade-off between in-domain accuracy and OOD robustness, and choosing the proper augmentations can help increase both in-domain accuracy and OOD robustness simultaneously.
* 7 pages, 4 figures
Machine-Learning Driven Drug Repurposing for COVID-19
Jun 25, 2020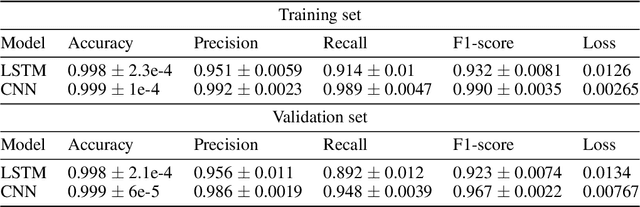
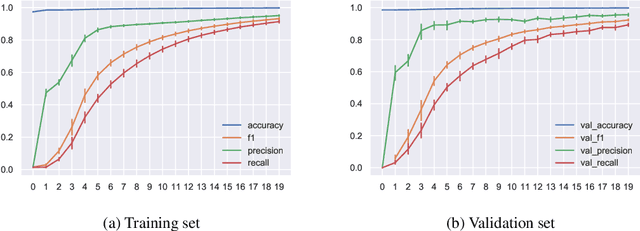
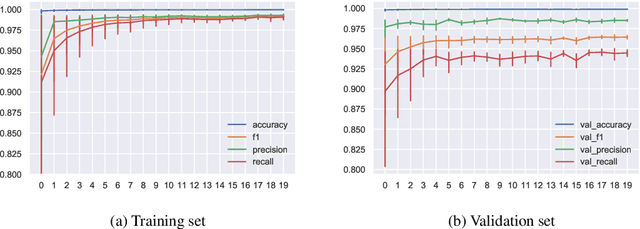
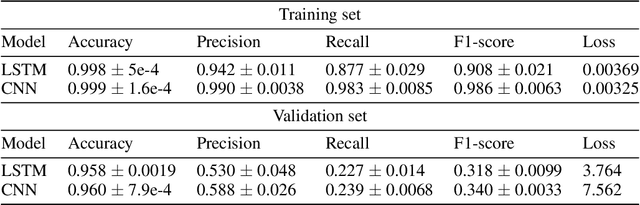
Abstract:The integration of machine learning methods into bioinformatics provides particular benefits in identifying how therapeutics effective in one context might have utility in an unknown clinical context or against a novel pathology. We aim to discover the underlying associations between viral proteins and antiviral therapeutics that are effective against them by employing neural network models. Using the National Center for Biotechnology Information virus protein database and the DrugVirus database, which provides a comprehensive report of broad-spectrum antiviral agents (BSAAs) and viruses they inhibit, we trained ANN models with virus protein sequences as inputs and antiviral agents deemed safe-in-humans as outputs. Model training excluded SARS-CoV-2 proteins and included only Phases II, III, IV and Approved level drugs. Using sequences for SARS-CoV-2 (the coronavirus that causes COVID-19) as inputs to the trained models produces outputs of tentative safe-in-human antiviral candidates for treating COVID-19. Our results suggest multiple drug candidates, some of which complement recent findings from noteworthy clinical studies. Our in-silico approach to drug repurposing has promise in identifying new drug candidates and treatments for other viruses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge