Scott Chapman
StomataSeg: Semi-Supervised Instance Segmentation for Sorghum Stomatal Components
Jan 31, 2026Abstract:Sorghum is a globally important cereal grown widely in water-limited and stress-prone regions. Its strong drought tolerance makes it a priority crop for climate-resilient agriculture. Improving water-use efficiency in sorghum requires precise characterisation of stomatal traits, as stomata control of gas exchange, transpiration and photosynthesis have a major influence on crop performance. Automated analysis of sorghum stomata is difficult because the stomata are small (often less than 40 $μ$m in length in grasses such as sorghum) and vary in shape across genotypes and leaf surfaces. Automated segmentation contributes to high-throughput stomatal phenotyping, yet current methods still face challenges related to nested small structures and annotation bottlenecks. In this paper, we propose a semi-supervised instance segmentation framework tailored for analysis of sorghum stomatal components. We collect and annotate a sorghum leaf imagery dataset containing 11,060 human-annotated patches, covering the three stomatal components (pore, guard cell and complex area) across multiple genotypes and leaf surfaces. To improve the detection of tiny structures, we split high-resolution microscopy images into overlapping small patches. We then apply a pseudo-labelling strategy to unannotated images, producing an additional 56,428 pseudo-labelled patches. Benchmarking across semantic and instance segmentation models shows substantial performance gains: for semantic models the top mIoU increases from 65.93% to 70.35%, whereas for instance models the top AP rises from 28.30% to 46.10%. These results demonstrate that combining patch-based preprocessing with semi-supervised learning significantly improves the segmentation of fine stomatal structures. The proposed framework supports scalable extraction of stomatal traits and facilitates broader adoption of AI-driven phenotyping in crop science.
FoMo4Wheat: Toward reliable crop vision foundation models with globally curated data
Sep 08, 2025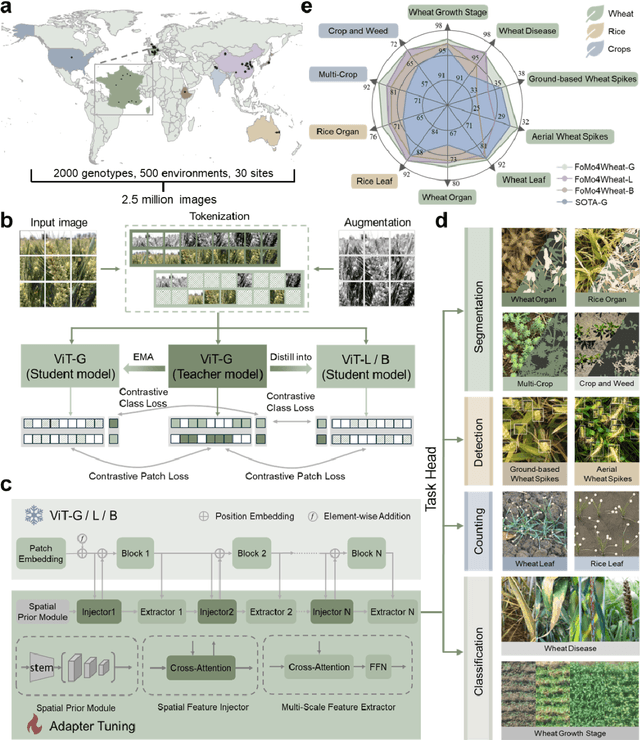
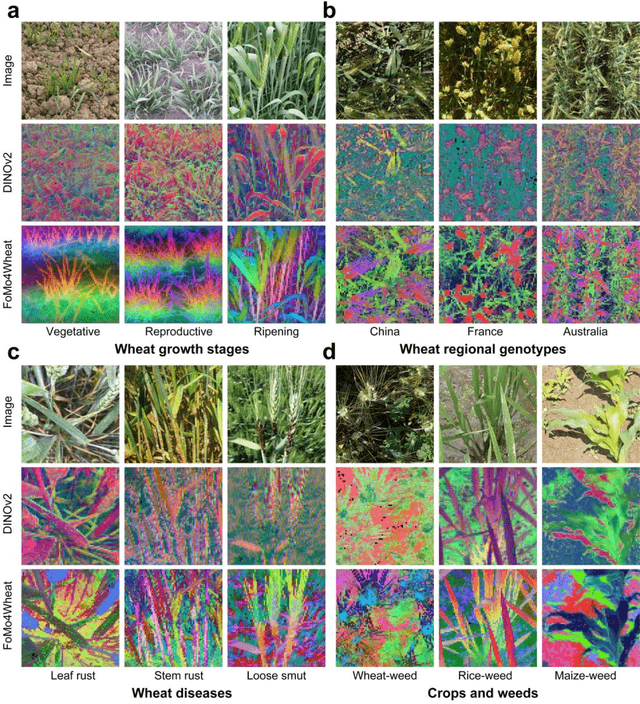
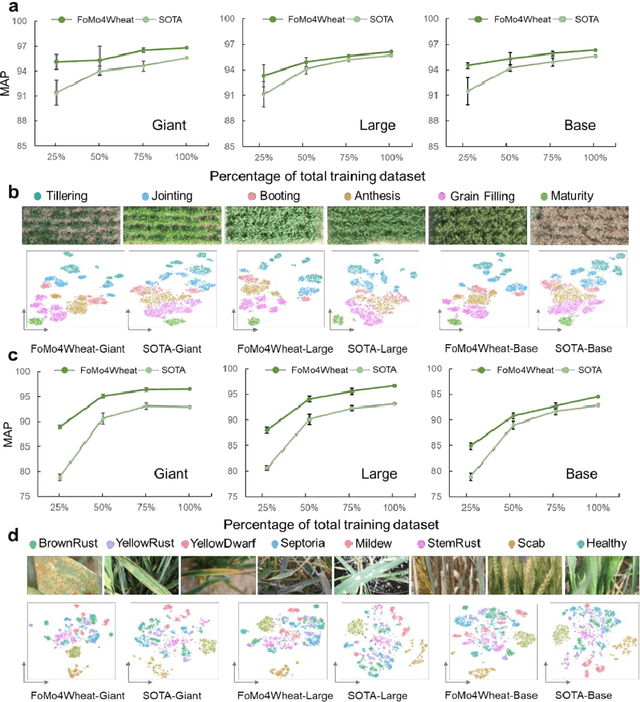
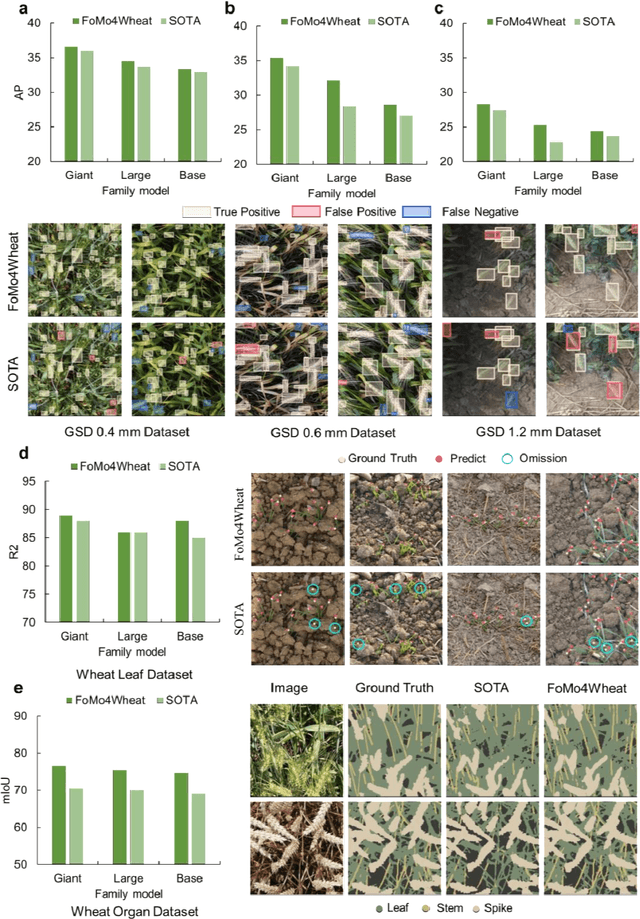
Abstract:Vision-driven field monitoring is central to digital agriculture, yet models built on general-domain pretrained backbones often fail to generalize across tasks, owing to the interaction of fine, variable canopy structures with fluctuating field conditions. We present FoMo4Wheat, one of the first crop-domain vision foundation model pretrained with self-supervision on ImAg4Wheat, the largest and most diverse wheat image dataset to date (2.5 million high-resolution images collected over a decade at 30 global sites, spanning >2,000 genotypes and >500 environmental conditions). This wheat-specific pretraining yields representations that are robust for wheat and transferable to other crops and weeds. Across ten in-field vision tasks at canopy and organ levels, FoMo4Wheat models consistently outperform state-of-the-art models pretrained on general-domain dataset. These results demonstrate the value of crop-specific foundation models for reliable in-field perception and chart a path toward a universal crop foundation model with cross-species and cross-task capabilities. FoMo4Wheat models and the ImAg4Wheat dataset are publicly available online: https://github.com/PheniX-Lab/FoMo4Wheat and https://huggingface.co/PheniX-Lab/FoMo4Wheat. The demonstration website is: https://fomo4wheat.phenix-lab.com/.
Track Any Peppers: Weakly Supervised Sweet Pepper Tracking Using VLMs
Nov 11, 2024Abstract:In the Detection and Multi-Object Tracking of Sweet Peppers Challenge, we present Track Any Peppers (TAP) - a weakly supervised ensemble technique for sweet peppers tracking. TAP leverages the zero-shot detection capabilities of vision-language foundation models like Grounding DINO to automatically generate pseudo-labels for sweet peppers in video sequences with minimal human intervention. These pseudo-labels, refined when necessary, are used to train a YOLOv8 segmentation network. To enhance detection accuracy under challenging conditions, we incorporate pre-processing techniques such as relighting adjustments and apply depth-based filtering during post-inference. For object tracking, we integrate the Matching by Segment Anything (MASA) adapter with the BoT-SORT algorithm. Our approach achieves a HOTA score of 80.4%, MOTA of 66.1%, Recall of 74.0%, and Precision of 90.7%, demonstrating effective tracking of sweet peppers without extensive manual effort. This work highlights the potential of foundation models for efficient and accurate object detection and tracking in agricultural settings.
CF-PRNet: Coarse-to-Fine Prototype Refining Network for Point Cloud Completion and Reconstruction
Sep 13, 2024



Abstract:In modern agriculture, precise monitoring of plants and fruits is crucial for tasks such as high-throughput phenotyping and automated harvesting. This paper addresses the challenge of reconstructing accurate 3D shapes of fruits from partial views, which is common in agricultural settings. We introduce CF-PRNet, a coarse-to-fine prototype refining network, leverages high-resolution 3D data during the training phase but requires only a single RGB-D image for real-time inference. Our approach begins by extracting the incomplete point cloud data that constructed from a partial view of a fruit with a series of convolutional blocks. The extracted features inform the generation of scaling vectors that refine two sequentially constructed 3D mesh prototypes - one coarse and one fine-grained. This progressive refinement facilitates the detailed completion of the final point clouds, achieving detailed and accurate reconstructions. CF-PRNet demonstrates excellent performance metrics with a Chamfer Distance of 3.78, an F1 Score of 66.76%, a Precision of 56.56%, and a Recall of 85.31%, and win the first place in the Shape Completion and Reconstruction of Sweet Peppers Challenge.
PlantSeg: A Large-Scale In-the-wild Dataset for Plant Disease Segmentation
Sep 06, 2024



Abstract:Plant diseases pose significant threats to agriculture. It necessitates proper diagnosis and effective treatment to safeguard crop yields. To automate the diagnosis process, image segmentation is usually adopted for precisely identifying diseased regions, thereby advancing precision agriculture. Developing robust image segmentation models for plant diseases demands high-quality annotations across numerous images. However, existing plant disease datasets typically lack segmentation labels and are often confined to controlled laboratory settings, which do not adequately reflect the complexity of natural environments. Motivated by this fact, we established PlantSeg, a large-scale segmentation dataset for plant diseases. PlantSeg distinguishes itself from existing datasets in three key aspects. (1) Annotation type: Unlike the majority of existing datasets that only contain class labels or bounding boxes, each image in PlantSeg includes detailed and high-quality segmentation masks, associated with plant types and disease names. (2) Image source: Unlike typical datasets that contain images from laboratory settings, PlantSeg primarily comprises in-the-wild plant disease images. This choice enhances the practical applicability, as the trained models can be applied for integrated disease management. (3) Scale: PlantSeg is extensive, featuring 11,400 images with disease segmentation masks and an additional 8,000 healthy plant images categorized by plant type. Extensive technical experiments validate the high quality of PlantSeg's annotations. This dataset not only allows researchers to evaluate their image classification methods but also provides a critical foundation for developing and benchmarking advanced plant disease segmentation algorithms.
Divide and Ensemble: Progressively Learning for the Unknown
Oct 09, 2023
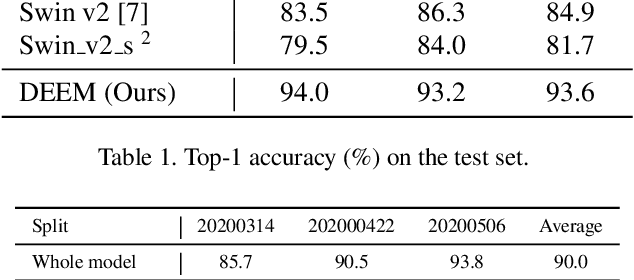


Abstract:In the wheat nutrient deficiencies classification challenge, we present the DividE and EnseMble (DEEM) method for progressive test data predictions. We find that (1) test images are provided in the challenge; (2) samples are equipped with their collection dates; (3) the samples of different dates show notable discrepancies. Based on the findings, we partition the dataset into discrete groups by the dates and train models on each divided group. We then adopt the pseudo-labeling approach to label the test data and incorporate those with high confidence into the training set. In pseudo-labeling, we leverage models ensemble with different architectures to enhance the reliability of predictions. The pseudo-labeling and ensembled model training are iteratively conducted until all test samples are labeled. Finally, the separated models for each group are unified to obtain the model for the whole dataset. Our method achieves an average of 93.6\% Top-1 test accuracy~(94.0\% on WW2020 and 93.2\% on WR2021) and wins the 1$st$ place in the Deep Nutrient Deficiency Challenge~\footnote{https://cvppa2023.github.io/challenges/}.
Global Wheat Head Dataset 2021: more diversity to improve the benchmarking of wheat head localization methods
Jun 03, 2021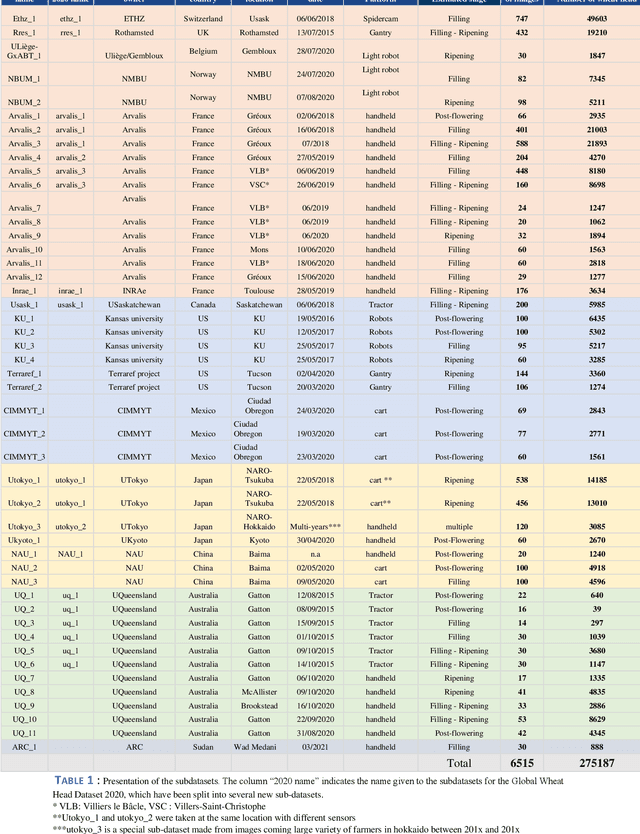
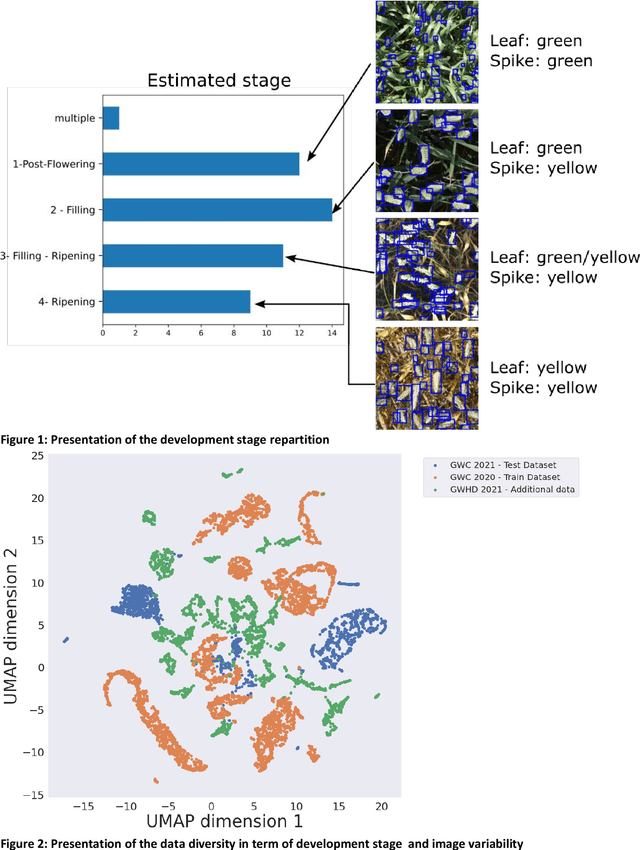
Abstract:The Global Wheat Head Detection (GWHD) dataset was created in 2020 and has assembled 193,634 labelled wheat heads from 4,700 RGB images acquired from various acquisition platforms and 7 countries/institutions. With an associated competition hosted in Kaggle, GWHD has successfully attracted attention from both the computer vision and agricultural science communities. From this first experience in 2020, a few avenues for improvements have been identified, especially from the perspective of data size, head diversity and label reliability. To address these issues, the 2020 dataset has been reexamined, relabeled, and augmented by adding 1,722 images from 5 additional countries, allowing for 81,553 additional wheat heads to be added. We now release a new version of the Global Wheat Head Detection (GWHD) dataset in 2021, which is bigger, more diverse, and less noisy than the 2020 version. The GWHD 2021 is now publicly available at http://www.global-wheat.com/ and a new data challenge has been organized on AIcrowd to make use of this updated dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge