Qingzheng Xu
MM-WLAuslan: Multi-View Multi-Modal Word-Level Australian Sign Language Recognition Dataset
Oct 25, 2024



Abstract:Isolated Sign Language Recognition (ISLR) focuses on identifying individual sign language glosses. Considering the diversity of sign languages across geographical regions, developing region-specific ISLR datasets is crucial for supporting communication and research. Auslan, as a sign language specific to Australia, still lacks a dedicated large-scale word-level dataset for the ISLR task. To fill this gap, we curate \underline{\textbf{the first}} large-scale Multi-view Multi-modal Word-Level Australian Sign Language recognition dataset, dubbed MM-WLAuslan. Compared to other publicly available datasets, MM-WLAuslan exhibits three significant advantages: (1) the largest amount of data, (2) the most extensive vocabulary, and (3) the most diverse of multi-modal camera views. Specifically, we record 282K+ sign videos covering 3,215 commonly used Auslan glosses presented by 73 signers in a studio environment. Moreover, our filming system includes two different types of cameras, i.e., three Kinect-V2 cameras and a RealSense camera. We position cameras hemispherically around the front half of the model and simultaneously record videos using all four cameras. Furthermore, we benchmark results with state-of-the-art methods for various multi-modal ISLR settings on MM-WLAuslan, including multi-view, cross-camera, and cross-view. Experiment results indicate that MM-WLAuslan is a challenging ISLR dataset, and we hope this dataset will contribute to the development of Auslan and the advancement of sign languages worldwide. All datasets and benchmarks are available at MM-WLAuslan.
Divide and Ensemble: Progressively Learning for the Unknown
Oct 09, 2023
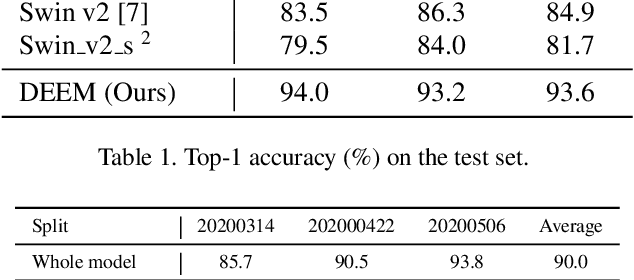


Abstract:In the wheat nutrient deficiencies classification challenge, we present the DividE and EnseMble (DEEM) method for progressive test data predictions. We find that (1) test images are provided in the challenge; (2) samples are equipped with their collection dates; (3) the samples of different dates show notable discrepancies. Based on the findings, we partition the dataset into discrete groups by the dates and train models on each divided group. We then adopt the pseudo-labeling approach to label the test data and incorporate those with high confidence into the training set. In pseudo-labeling, we leverage models ensemble with different architectures to enhance the reliability of predictions. The pseudo-labeling and ensembled model training are iteratively conducted until all test samples are labeled. Finally, the separated models for each group are unified to obtain the model for the whole dataset. Our method achieves an average of 93.6\% Top-1 test accuracy~(94.0\% on WW2020 and 93.2\% on WR2021) and wins the 1$st$ place in the Deep Nutrient Deficiency Challenge~\footnote{https://cvppa2023.github.io/challenges/}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge