Tianqi Wei
RAP: Runtime-Adaptive Pruning for LLM Inference
May 26, 2025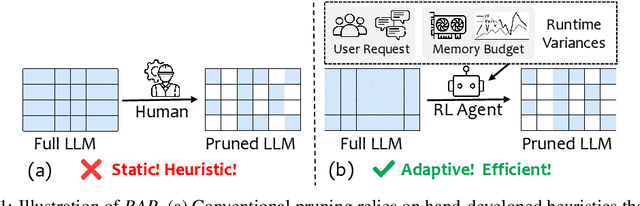

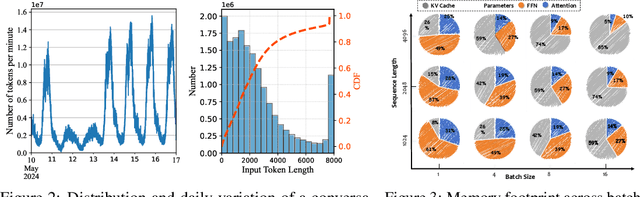

Abstract:Large language models (LLMs) excel at language understanding and generation, but their enormous computational and memory requirements hinder deployment. Compression offers a potential solution to mitigate these constraints. However, most existing methods rely on fixed heuristics and thus fail to adapt to runtime memory variations or heterogeneous KV-cache demands arising from diverse user requests. To address these limitations, we propose RAP, an elastic pruning framework driven by reinforcement learning (RL) that dynamically adjusts compression strategies in a runtime-aware manner. Specifically, RAP dynamically tracks the evolving ratio between model parameters and KV-cache across practical execution. Recognizing that FFNs house most parameters, whereas parameter -light attention layers dominate KV-cache formation, the RL agent retains only those components that maximize utility within the current memory budget, conditioned on instantaneous workload and device state. Extensive experiments results demonstrate that RAP outperforms state-of-the-art baselines, marking the first time to jointly consider model weights and KV-cache on the fly.
Track Any Peppers: Weakly Supervised Sweet Pepper Tracking Using VLMs
Nov 11, 2024Abstract:In the Detection and Multi-Object Tracking of Sweet Peppers Challenge, we present Track Any Peppers (TAP) - a weakly supervised ensemble technique for sweet peppers tracking. TAP leverages the zero-shot detection capabilities of vision-language foundation models like Grounding DINO to automatically generate pseudo-labels for sweet peppers in video sequences with minimal human intervention. These pseudo-labels, refined when necessary, are used to train a YOLOv8 segmentation network. To enhance detection accuracy under challenging conditions, we incorporate pre-processing techniques such as relighting adjustments and apply depth-based filtering during post-inference. For object tracking, we integrate the Matching by Segment Anything (MASA) adapter with the BoT-SORT algorithm. Our approach achieves a HOTA score of 80.4%, MOTA of 66.1%, Recall of 74.0%, and Precision of 90.7%, demonstrating effective tracking of sweet peppers without extensive manual effort. This work highlights the potential of foundation models for efficient and accurate object detection and tracking in agricultural settings.
CF-PRNet: Coarse-to-Fine Prototype Refining Network for Point Cloud Completion and Reconstruction
Sep 13, 2024



Abstract:In modern agriculture, precise monitoring of plants and fruits is crucial for tasks such as high-throughput phenotyping and automated harvesting. This paper addresses the challenge of reconstructing accurate 3D shapes of fruits from partial views, which is common in agricultural settings. We introduce CF-PRNet, a coarse-to-fine prototype refining network, leverages high-resolution 3D data during the training phase but requires only a single RGB-D image for real-time inference. Our approach begins by extracting the incomplete point cloud data that constructed from a partial view of a fruit with a series of convolutional blocks. The extracted features inform the generation of scaling vectors that refine two sequentially constructed 3D mesh prototypes - one coarse and one fine-grained. This progressive refinement facilitates the detailed completion of the final point clouds, achieving detailed and accurate reconstructions. CF-PRNet demonstrates excellent performance metrics with a Chamfer Distance of 3.78, an F1 Score of 66.76%, a Precision of 56.56%, and a Recall of 85.31%, and win the first place in the Shape Completion and Reconstruction of Sweet Peppers Challenge.
PlantSeg: A Large-Scale In-the-wild Dataset for Plant Disease Segmentation
Sep 06, 2024



Abstract:Plant diseases pose significant threats to agriculture. It necessitates proper diagnosis and effective treatment to safeguard crop yields. To automate the diagnosis process, image segmentation is usually adopted for precisely identifying diseased regions, thereby advancing precision agriculture. Developing robust image segmentation models for plant diseases demands high-quality annotations across numerous images. However, existing plant disease datasets typically lack segmentation labels and are often confined to controlled laboratory settings, which do not adequately reflect the complexity of natural environments. Motivated by this fact, we established PlantSeg, a large-scale segmentation dataset for plant diseases. PlantSeg distinguishes itself from existing datasets in three key aspects. (1) Annotation type: Unlike the majority of existing datasets that only contain class labels or bounding boxes, each image in PlantSeg includes detailed and high-quality segmentation masks, associated with plant types and disease names. (2) Image source: Unlike typical datasets that contain images from laboratory settings, PlantSeg primarily comprises in-the-wild plant disease images. This choice enhances the practical applicability, as the trained models can be applied for integrated disease management. (3) Scale: PlantSeg is extensive, featuring 11,400 images with disease segmentation masks and an additional 8,000 healthy plant images categorized by plant type. Extensive technical experiments validate the high quality of PlantSeg's annotations. This dataset not only allows researchers to evaluate their image classification methods but also provides a critical foundation for developing and benchmarking advanced plant disease segmentation algorithms.
Snap and Diagnose: An Advanced Multimodal Retrieval System for Identifying Plant Diseases in the Wild
Aug 27, 2024


Abstract:Plant disease recognition is a critical task that ensures crop health and mitigates the damage caused by diseases. A handy tool that enables farmers to receive a diagnosis based on query pictures or the text description of suspicious plants is in high demand for initiating treatment before potential diseases spread further. In this paper, we develop a multimodal plant disease image retrieval system to support disease search based on either image or text prompts. Specifically, we utilize the largest in-the-wild plant disease dataset PlantWild, which includes over 18,000 images across 89 categories, to provide a comprehensive view of potential diseases relating to the query. Furthermore, cross-modal retrieval is achieved in the developed system, facilitated by a novel CLIP-based vision-language model that encodes both disease descriptions and disease images into the same latent space. Built on top of the retriever, our retrieval system allows users to upload either plant disease images or disease descriptions to retrieve the corresponding images with similar characteristics from the disease dataset to suggest candidate diseases for end users' consideration.
Benchmarking In-the-wild Multimodal Disease Recognition and A Versatile Baseline
Aug 06, 2024Abstract:Existing plant disease classification models have achieved remarkable performance in recognizing in-laboratory diseased images. However, their performance often significantly degrades in classifying in-the-wild images. Furthermore, we observed that in-the-wild plant images may exhibit similar appearances across various diseases (i.e., small inter-class discrepancy) while the same diseases may look quite different (i.e., large intra-class variance). Motivated by this observation, we propose an in-the-wild multimodal plant disease recognition dataset that contains the largest number of disease classes but also text-based descriptions for each disease. Particularly, the newly provided text descriptions are introduced to provide rich information in textual modality and facilitate in-the-wild disease classification with small inter-class discrepancy and large intra-class variance issues. Therefore, our proposed dataset can be regarded as an ideal testbed for evaluating disease recognition methods in the real world. In addition, we further present a strong yet versatile baseline that models text descriptions and visual data through multiple prototypes for a given class. By fusing the contributions of multimodal prototypes in classification, our baseline can effectively address the small inter-class discrepancy and large intra-class variance issues. Remarkably, our baseline model can not only classify diseases but also recognize diseases in few-shot or training-free scenarios. Extensive benchmarking results demonstrate that our proposed in-the-wild multimodal dataset sets many new challenges to the plant disease recognition task and there is a large space to improve for future works.
Learning Quadruped Locomotion using Bio-Inspired Neural Networks with Intrinsic Rhythmicity
May 12, 2023
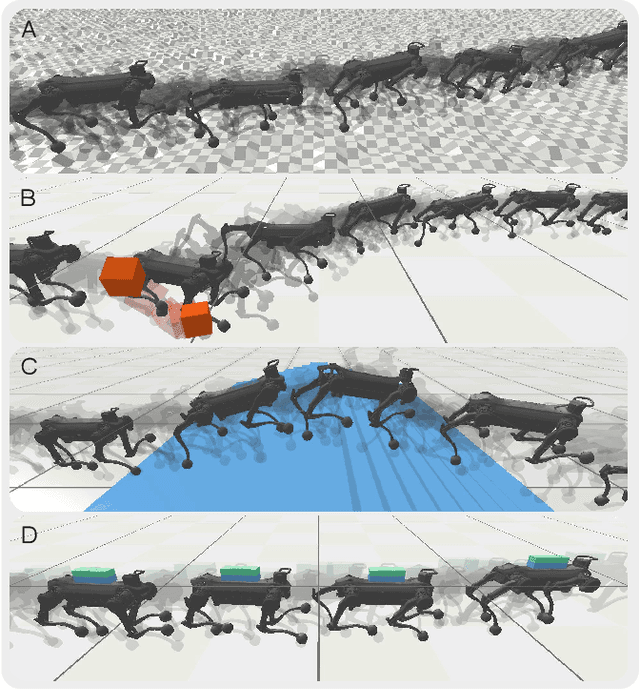
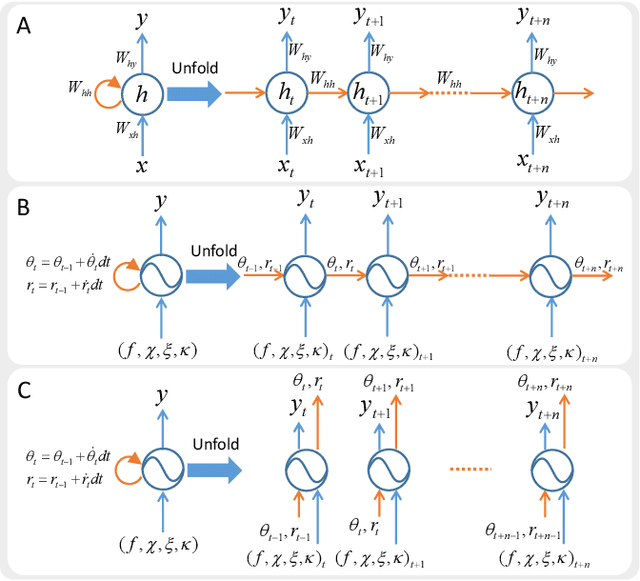
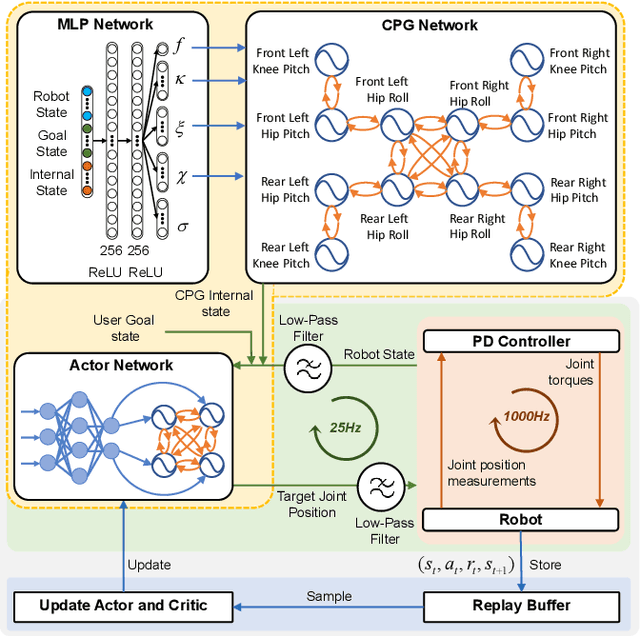
Abstract:Biological studies reveal that neural circuits located at the spinal cord called central pattern generator (CPG) oscillates and generates rhythmic signals, which are the underlying mechanism responsible for rhythmic locomotion behaviors of animals. Inspired by CPG's capability to naturally generate rhythmic patterns, researchers have attempted to create mathematical models of CPG and utilize them for the locomotion of legged robots. In this paper, we propose a network architecture that incorporates CPGs for rhythmic pattern generation and a multi-layer perceptron (MLP) network for sensory feedback. We also proposed a method that reformulates CPGs into a fully-differentiable stateless network, allowing CPGs and MLP to be jointly trained with gradient-based learning. The results show that our proposed method learned agile and dynamic locomotion policies which are capable of blind traversal over uneven terrain and resist external pushes. Simulation results also show that the learned policies are capable of self-modulating step frequency and step length to adapt to the locomotion velocity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge