Sayed Hadi Hashemi
Gemini 1.5: Unlocking multimodal understanding across millions of tokens of context
Mar 08, 2024Abstract:In this report, we present the latest model of the Gemini family, Gemini 1.5 Pro, a highly compute-efficient multimodal mixture-of-experts model capable of recalling and reasoning over fine-grained information from millions of tokens of context, including multiple long documents and hours of video and audio. Gemini 1.5 Pro achieves near-perfect recall on long-context retrieval tasks across modalities, improves the state-of-the-art in long-document QA, long-video QA and long-context ASR, and matches or surpasses Gemini 1.0 Ultra's state-of-the-art performance across a broad set of benchmarks. Studying the limits of Gemini 1.5 Pro's long-context ability, we find continued improvement in next-token prediction and near-perfect retrieval (>99%) up to at least 10M tokens, a generational leap over existing models such as Claude 2.1 (200k) and GPT-4 Turbo (128k). Finally, we highlight surprising new capabilities of large language models at the frontier; when given a grammar manual for Kalamang, a language with fewer than 200 speakers worldwide, the model learns to translate English to Kalamang at a similar level to a person who learned from the same content.
Gemini: A Family of Highly Capable Multimodal Models
Dec 19, 2023Abstract:This report introduces a new family of multimodal models, Gemini, that exhibit remarkable capabilities across image, audio, video, and text understanding. The Gemini family consists of Ultra, Pro, and Nano sizes, suitable for applications ranging from complex reasoning tasks to on-device memory-constrained use-cases. Evaluation on a broad range of benchmarks shows that our most-capable Gemini Ultra model advances the state of the art in 30 of 32 of these benchmarks - notably being the first model to achieve human-expert performance on the well-studied exam benchmark MMLU, and improving the state of the art in every one of the 20 multimodal benchmarks we examined. We believe that the new capabilities of Gemini models in cross-modal reasoning and language understanding will enable a wide variety of use cases and we discuss our approach toward deploying them responsibly to users.
GenoML: Automated Machine Learning for Genomics
Mar 04, 2021Abstract:GenoML is a Python package automating machine learning workflows for genomics (genetics and multi-omics) with an open science philosophy. Genomics data require significant domain expertise to clean, pre-process, harmonize and perform quality control of the data. Furthermore, tuning, validation, and interpretation involve taking into account the biology and possibly the limitations of the underlying data collection, protocols, and technology. GenoML's mission is to bring machine learning for genomics and clinical data to non-experts by developing an easy-to-use tool that automates the full development, evaluation, and deployment process. Emphasis is put on open science to make workflows easily accessible, replicable, and transferable within the scientific community. Source code and documentation is available at https://genoml.com.
Caramel: Accelerating Decentralized Distributed Deep Learning with Computation Scheduling
Apr 29, 2020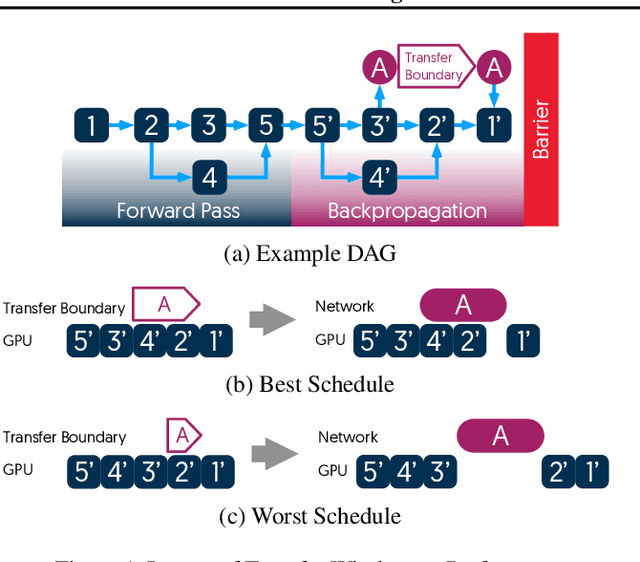
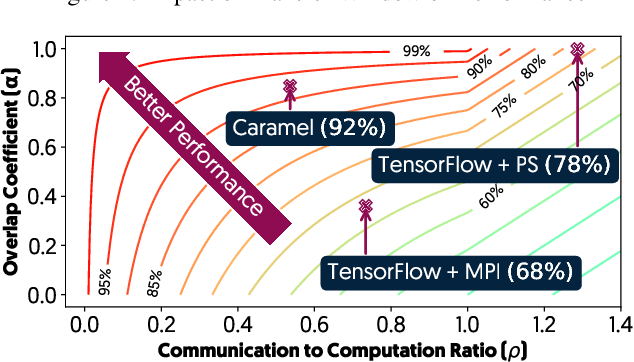
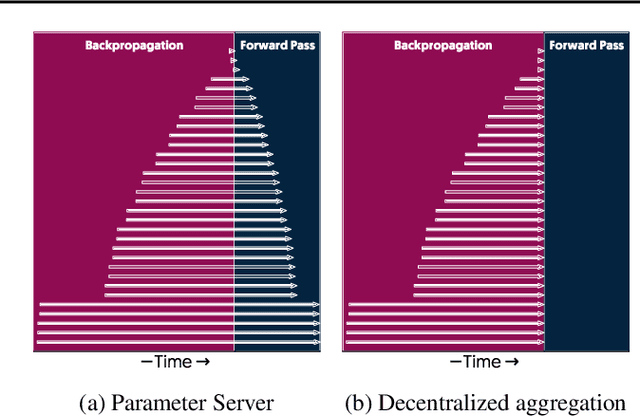
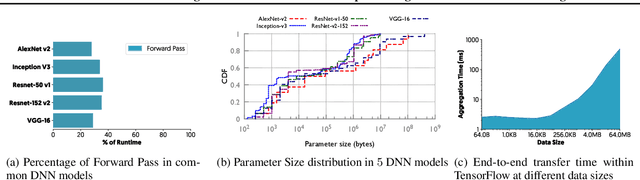
Abstract:The method of choice for parameter aggregation in Deep Neural Network (DNN) training, a network-intensive task, is shifting from the Parameter Server model to decentralized aggregation schemes (AllReduce) inspired by theoretical guarantees of better performance. However, current implementations of AllReduce overlook the interdependence of communication and computation, resulting in significant performance degradation. In this paper, we develop Caramel, a system that accelerates decentralized distributed deep learning through model-aware computation scheduling and communication optimizations for AllReduce. Caramel achieves this goal through (a) computation DAG scheduling that expands the feasible window of transfer for each parameter (transfer boundaries), and (b) network optimizations for smoothening of the load including adaptive batching and pipelining of parameter transfers. Caramel maintains the correctness of the dataflow model, is hardware-independent, and does not require any user-level or framework-level changes. We implement Caramel over TensorFlow and show that the iteration time of DNN training can be improved by up to 3.62x in a cloud environment.
TicTac: Accelerating Distributed Deep Learning with Communication Scheduling
Oct 04, 2018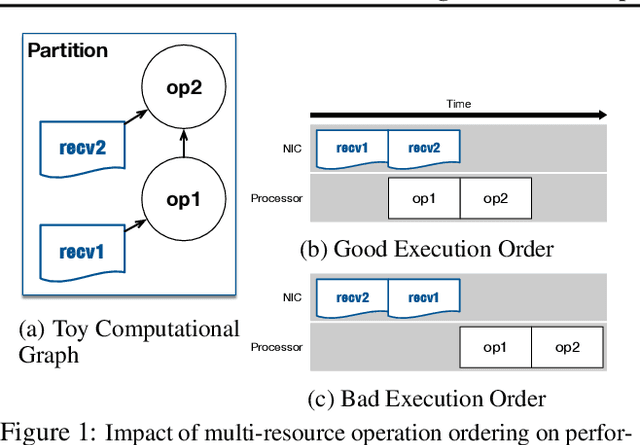
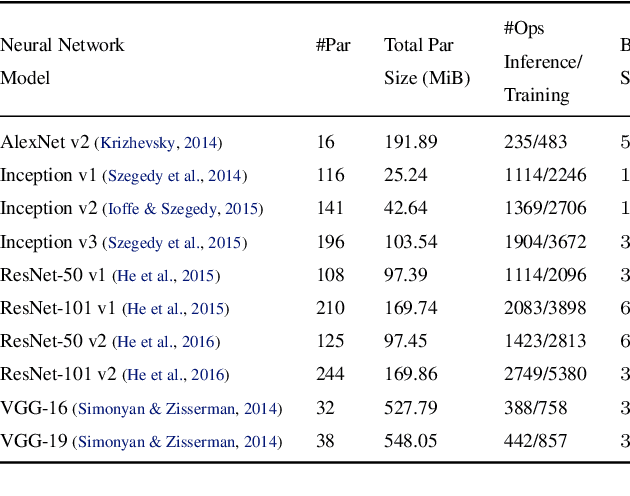
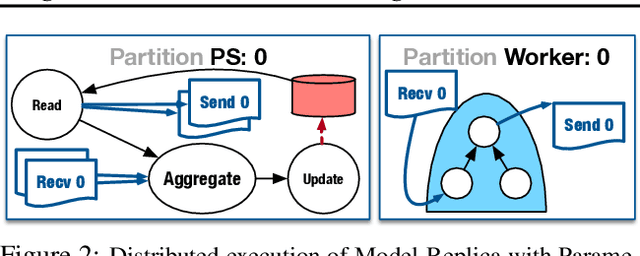
Abstract:State-of-the-art deep learning systems rely on iterative distributed training to tackle the increasing complexity of models and input data. The iteration time in these communication-heavy systems depends on the computation time, communication time and the extent of overlap of computation and communication. In this work, we identify a shortcoming in systems with graph representation for computation, such as TensorFlow and PyTorch, that result in high variance in iteration time --- random order of received parameters across workers. We develop a system, TicTac, to improve the iteration time by fixing this issue in distributed deep learning with Parameter Servers while guaranteeing near-optimal overlap of communication and computation. TicTac identifies and enforces an order of network transfers which improves the iteration time using prioritization. Our system is implemented over TensorFlow and requires no changes to the model or developer inputs. TicTac improves the throughput by up to $37.7\%$ in inference and $19.2\%$ in training, while also reducing straggler effect by up to $2.3\times$. Our code is publicly available.
Toward Scalable Machine Learning and Data Mining: the Bioinformatics Case
Sep 29, 2017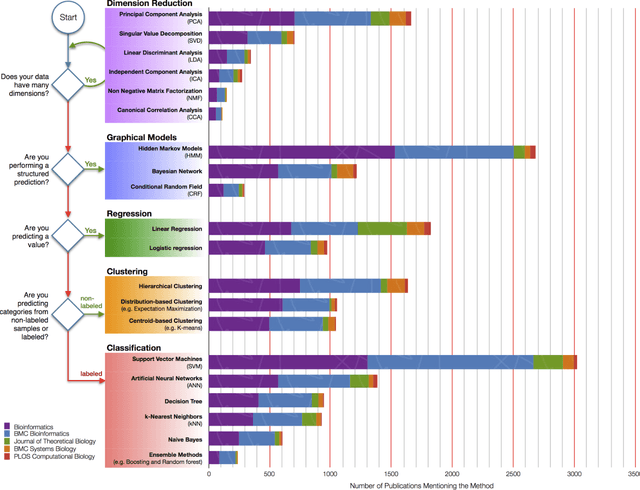
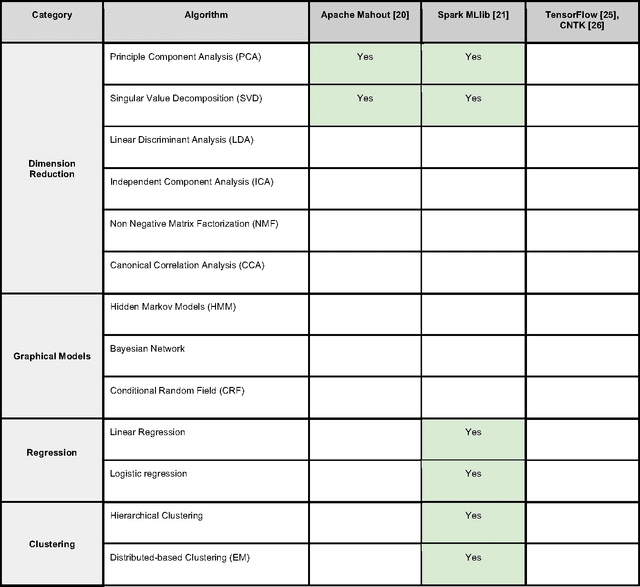
Abstract:In an effort to overcome the data deluge in computational biology and bioinformatics and to facilitate bioinformatics research in the era of big data, we identify some of the most influential algorithms that have been widely used in the bioinformatics community. These top data mining and machine learning algorithms cover classification, clustering, regression, graphical model-based learning, and dimensionality reduction. The goal of this study is to guide the focus of scalable computing experts in the endeavor of applying new storage and scalable computation designs to bioinformatics algorithms that merit their attention most, following the engineering maxim of "optimize the common case".
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge