Roy H. Campbell
GenoML: Automated Machine Learning for Genomics
Mar 04, 2021Abstract:GenoML is a Python package automating machine learning workflows for genomics (genetics and multi-omics) with an open science philosophy. Genomics data require significant domain expertise to clean, pre-process, harmonize and perform quality control of the data. Furthermore, tuning, validation, and interpretation involve taking into account the biology and possibly the limitations of the underlying data collection, protocols, and technology. GenoML's mission is to bring machine learning for genomics and clinical data to non-experts by developing an easy-to-use tool that automates the full development, evaluation, and deployment process. Emphasis is put on open science to make workflows easily accessible, replicable, and transferable within the scientific community. Source code and documentation is available at https://genoml.com.
TicTac: Accelerating Distributed Deep Learning with Communication Scheduling
Oct 04, 2018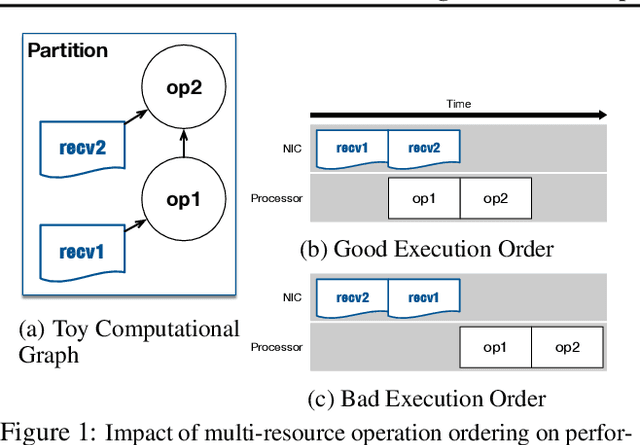
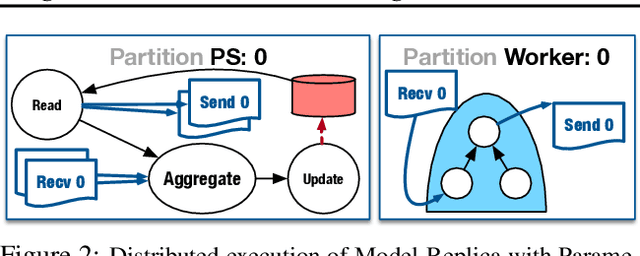
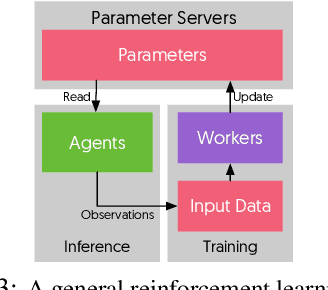
Abstract:State-of-the-art deep learning systems rely on iterative distributed training to tackle the increasing complexity of models and input data. The iteration time in these communication-heavy systems depends on the computation time, communication time and the extent of overlap of computation and communication. In this work, we identify a shortcoming in systems with graph representation for computation, such as TensorFlow and PyTorch, that result in high variance in iteration time --- random order of received parameters across workers. We develop a system, TicTac, to improve the iteration time by fixing this issue in distributed deep learning with Parameter Servers while guaranteeing near-optimal overlap of communication and computation. TicTac identifies and enforces an order of network transfers which improves the iteration time using prioritization. Our system is implemented over TensorFlow and requires no changes to the model or developer inputs. TicTac improves the throughput by up to $37.7\%$ in inference and $19.2\%$ in training, while also reducing straggler effect by up to $2.3\times$. Our code is publicly available.
Stochastic Variational Video Prediction
Mar 06, 2018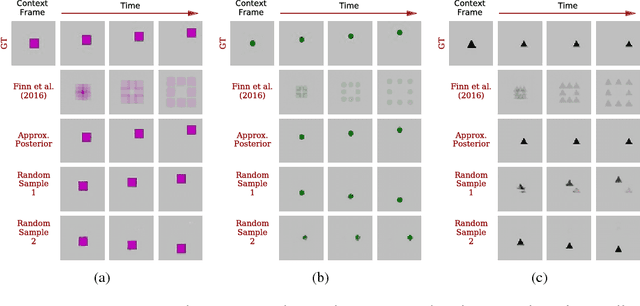
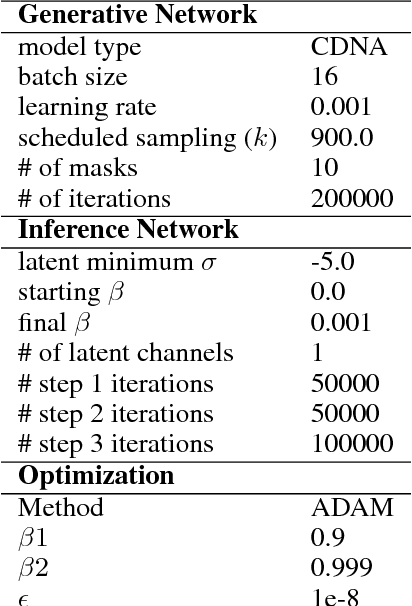
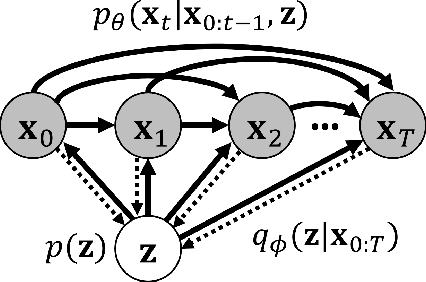
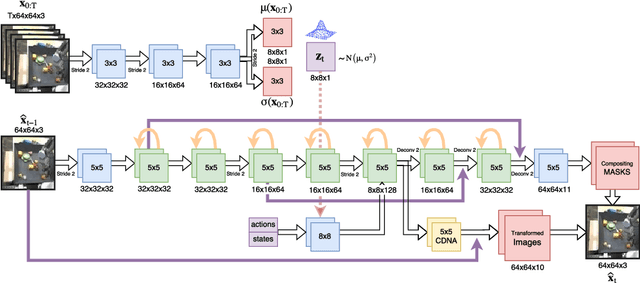
Abstract:Predicting the future in real-world settings, particularly from raw sensory observations such as images, is exceptionally challenging. Real-world events can be stochastic and unpredictable, and the high dimensionality and complexity of natural images requires the predictive model to build an intricate understanding of the natural world. Many existing methods tackle this problem by making simplifying assumptions about the environment. One common assumption is that the outcome is deterministic and there is only one plausible future. This can lead to low-quality predictions in real-world settings with stochastic dynamics. In this paper, we develop a stochastic variational video prediction (SV2P) method that predicts a different possible future for each sample of its latent variables. To the best of our knowledge, our model is the first to provide effective stochastic multi-frame prediction for real-world video. We demonstrate the capability of the proposed method in predicting detailed future frames of videos on multiple real-world datasets, both action-free and action-conditioned. We find that our proposed method produces substantially improved video predictions when compared to the same model without stochasticity, and to other stochastic video prediction methods. Our SV2P implementation will be open sourced upon publication.
Toward Scalable Machine Learning and Data Mining: the Bioinformatics Case
Sep 29, 2017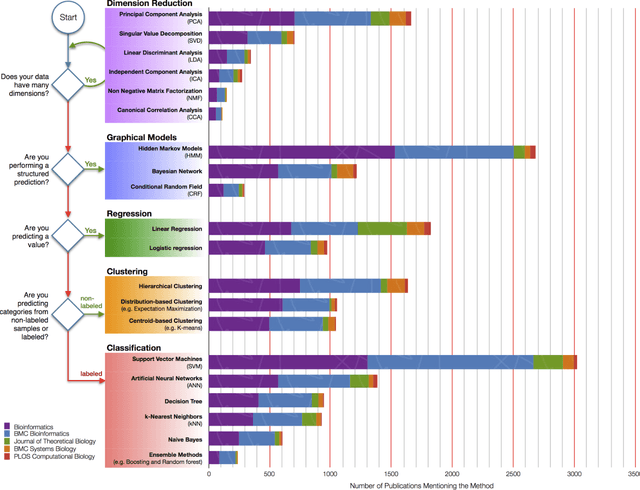
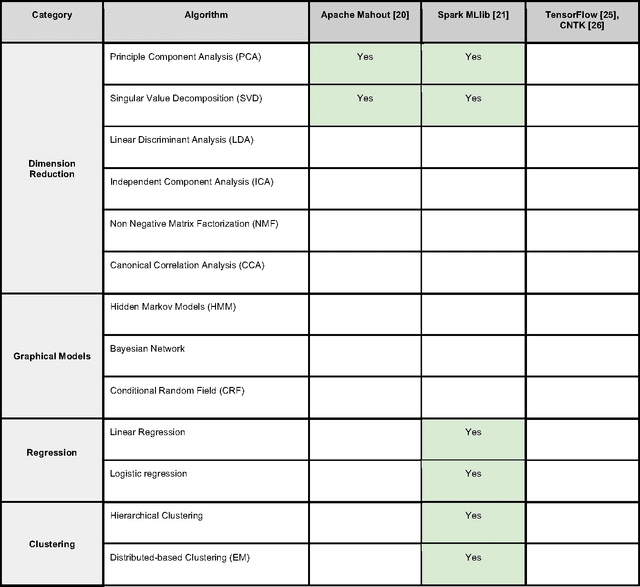
Abstract:In an effort to overcome the data deluge in computational biology and bioinformatics and to facilitate bioinformatics research in the era of big data, we identify some of the most influential algorithms that have been widely used in the bioinformatics community. These top data mining and machine learning algorithms cover classification, clustering, regression, graphical model-based learning, and dimensionality reduction. The goal of this study is to guide the focus of scalable computing experts in the endeavor of applying new storage and scalable computation designs to bioinformatics algorithms that merit their attention most, following the engineering maxim of "optimize the common case".
Fast Generation for Convolutional Autoregressive Models
Apr 20, 2017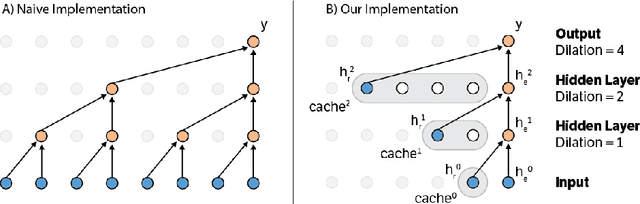
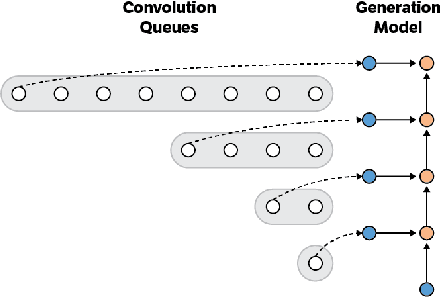
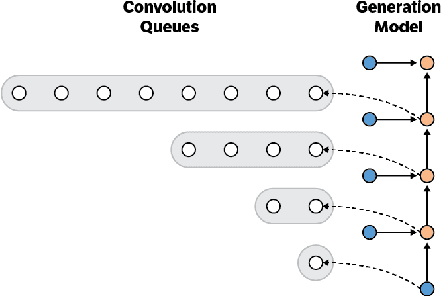

Abstract:Convolutional autoregressive models have recently demonstrated state-of-the-art performance on a number of generation tasks. While fast, parallel training methods have been crucial for their success, generation is typically implemented in a na\"{i}ve fashion where redundant computations are unnecessarily repeated. This results in slow generation, making such models infeasible for production environments. In this work, we describe a method to speed up generation in convolutional autoregressive models. The key idea is to cache hidden states to avoid redundant computation. We apply our fast generation method to the Wavenet and PixelCNN++ models and achieve up to $21\times$ and $183\times$ speedups respectively.
NoiseOut: A Simple Way to Prune Neural Networks
Nov 18, 2016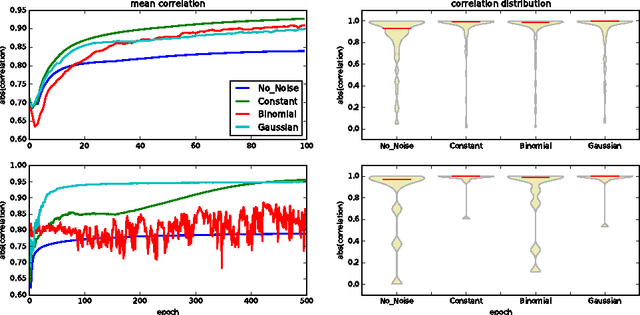
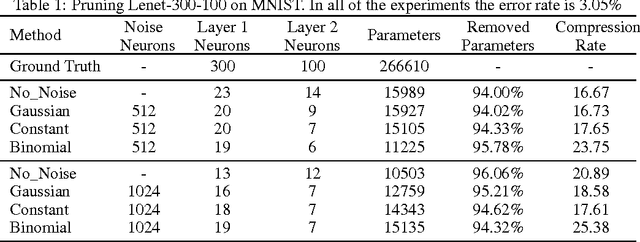
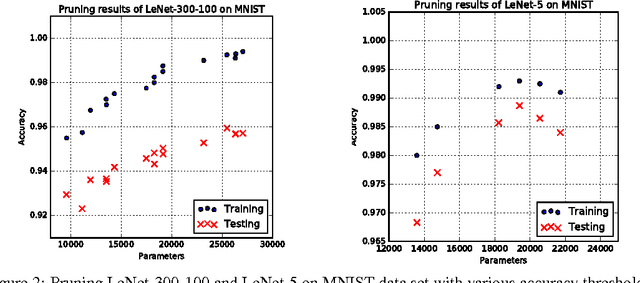
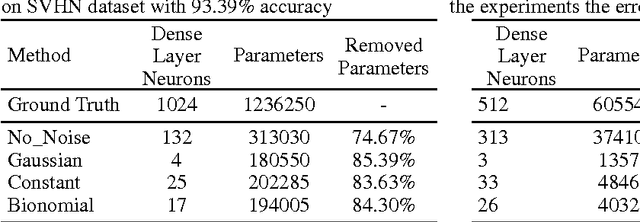
Abstract:Neural networks are usually over-parameterized with significant redundancy in the number of required neurons which results in unnecessary computation and memory usage at inference time. One common approach to address this issue is to prune these big networks by removing extra neurons and parameters while maintaining the accuracy. In this paper, we propose NoiseOut, a fully automated pruning algorithm based on the correlation between activations of neurons in the hidden layers. We prove that adding additional output neurons with entirely random targets results into a higher correlation between neurons which makes pruning by NoiseOut even more efficient. Finally, we test our method on various networks and datasets. These experiments exhibit high pruning rates while maintaining the accuracy of the original network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge