Satoshi Iizuka
Detection of Medial Epicondyle Avulsion in Elbow Ultrasound Images via Bone Structure Reconstruction
Jul 27, 2025Abstract:This study proposes a reconstruction-based framework for detecting medial epicondyle avulsion in elbow ultrasound images, trained exclusively on normal cases. Medial epicondyle avulsion, commonly observed in baseball players, involves bone detachment and deformity, often appearing as discontinuities in bone contour. Therefore, learning the structure and continuity of normal bone is essential for detecting such abnormalities. To achieve this, we propose a masked autoencoder-based, structure-aware reconstruction framework that learns the continuity of normal bone structures. Even in the presence of avulsion, the model attempts to reconstruct the normal structure, resulting in large reconstruction errors at the avulsion site. For evaluation, we constructed a novel dataset comprising normal and avulsion ultrasound images from 16 baseball players, with pixel-level annotations under orthopedic supervision. Our method outperformed existing approaches, achieving a pixel-wise AUC of 0.965 and an image-wise AUC of 0.967. The dataset is publicly available at: https://github.com/Akahori000/Ultrasound-Medial-Epicondyle-Avulsion-Dataset.
Measurement of Medial Elbow Joint Space using Landmark Detection
Dec 17, 2024Abstract:Ultrasound imaging of the medial elbow is crucial for the early identification of Ulnar Collateral Ligament (UCL) injuries. Specifically, measuring the elbow joint space in ultrasound images is used to assess the valgus instability of elbow. To automate this measurement, a precisely annotated dataset is necessary; however, no publicly available dataset has been proposed thus far. This study introduces a novel ultrasound medial elbow dataset for measuring joint space to diagnose Ulnar Collateral Ligament (UCL) injuries. The dataset comprises 4,201 medial elbow ultrasound images from 22 subjects, with landmark annotations on the humerus and ulna. The annotations are made precisely by the authors under the supervision of three orthopedic surgeons. We evaluated joint space measurement methods using our proposed dataset with several landmark detection approaches, including ViTPose, HRNet, PCT, YOLOv8, and U-Net. In addition, we propose using Shape Subspace (SS) for landmark refinement in heatmap-based landmark detection. The results show that the mean Euclidean distance error of joint space is 0.116 mm when using HRNet. Furthermore, the SS landmark refinement improves the mean absolute error of landmark positions by 0.010 mm with HRNet and by 0.103 mm with ViTPose on average. These highlight the potential for high-precision, real-time diagnosis of UCL injuries and associated risks, which could be leveraged in large-scale screening. Lastly, we demonstrate point-based segmentation of the humerus and ulna using the detected landmarks as input. The dataset will be made publicly available upon acceptance of this paper at: https://github.com/Akahori000/Ultrasound-Medial-Elbow-Dataset.
Point Cloud Novelty Detection Based on Latent Representations of a General Feature Extractor
Oct 13, 2024Abstract:We propose an effective unsupervised 3D point cloud novelty detection approach, leveraging a general point cloud feature extractor and a one-class classifier. The general feature extractor consists of a graph-based autoencoder and is trained once on a point cloud dataset such as a mathematically generated fractal 3D point cloud dataset that is independent of normal/abnormal categories. The input point clouds are first converted into latent vectors by the general feature extractor, and then one-class classification is performed on the latent vectors. Compared to existing methods measuring the reconstruction error in 3D coordinate space, our approach utilizes latent representations where the shape information is condensed, which allows more direct and effective novelty detection. We confirm that our general feature extractor can extract shape features of unseen categories, eliminating the need for autoencoder re-training and reducing the computational burden. We validate the performance of our method through experiments on several subsets of the ShapeNet dataset and demonstrate that our latent-based approach outperforms the existing methods.
Visual Grounding of Whole Radiology Reports for 3D CT Images
Dec 08, 2023Abstract:Building a large-scale training dataset is an essential problem in the development of medical image recognition systems. Visual grounding techniques, which automatically associate objects in images with corresponding descriptions, can facilitate labeling of large number of images. However, visual grounding of radiology reports for CT images remains challenging, because so many kinds of anomalies are detectable via CT imaging, and resulting report descriptions are long and complex. In this paper, we present the first visual grounding framework designed for CT image and report pairs covering various body parts and diverse anomaly types. Our framework combines two components of 1) anatomical segmentation of images, and 2) report structuring. The anatomical segmentation provides multiple organ masks of given CT images, and helps the grounding model recognize detailed anatomies. The report structuring helps to accurately extract information regarding the presence, location, and type of each anomaly described in corresponding reports. Given the two additional image/report features, the grounding model can achieve better localization. In the verification process, we constructed a large-scale dataset with region-description correspondence annotations for 10,410 studies of 7,321 unique patients. We evaluated our framework using grounding accuracy, the percentage of correctly localized anomalies, as a metric and demonstrated that the combination of the anatomical segmentation and the report structuring improves the performance with a large margin over the baseline model (66.0% vs 77.8%). Comparison with the prior techniques also showed higher performance of our method.
* 14 pages, 7 figures. Accepted at MICCAI 2023
Image Synthesis-based Late Stage Cancer Augmentation and Semi-Supervised Segmentation for MRI Rectal Cancer Staging
Dec 08, 2023Abstract:Rectal cancer is one of the most common diseases and a major cause of mortality. For deciding rectal cancer treatment plans, T-staging is important. However, evaluating the index from preoperative MRI images requires high radiologists' skill and experience. Therefore, the aim of this study is to segment the mesorectum, rectum, and rectal cancer region so that the system can predict T-stage from segmentation results. Generally, shortage of large and diverse dataset and high quality annotation are known to be the bottlenecks in computer aided diagnostics development. Regarding rectal cancer, advanced cancer images are very rare, and per-pixel annotation requires high radiologists' skill and time. Therefore, it is not feasible to collect comprehensive disease patterns in a training dataset. To tackle this, we propose two kinds of approaches of image synthesis-based late stage cancer augmentation and semi-supervised learning which is designed for T-stage prediction. In the image synthesis data augmentation approach, we generated advanced cancer images from labels. The real cancer labels were deformed to resemble advanced cancer labels by artificial cancer progress simulation. Next, we introduce a T-staging loss which enables us to train segmentation models from per-image T-stage labels. The loss works to keep inclusion/invasion relationships between rectum and cancer region consistent to the ground truth T-stage. The verification tests show that the proposed method obtains the best sensitivity (0.76) and specificity (0.80) in distinguishing between over T3 stage and underT2. In the ablation studies, our semi-supervised learning approach with the T-staging loss improved specificity by 0.13. Adding the image synthesis-based data augmentation improved the DICE score of invasion cancer area by 0.08 from baseline.
Diffusion-based Holistic Texture Rectification and Synthesis
Sep 26, 2023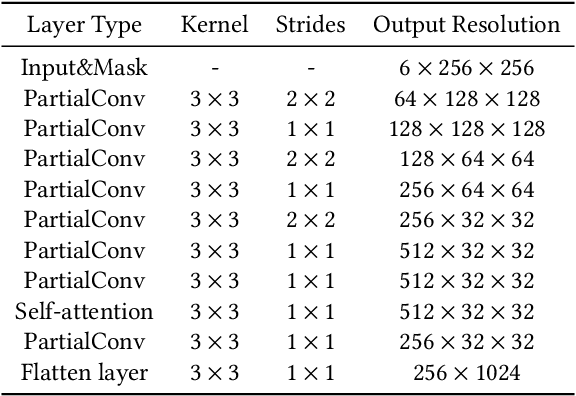
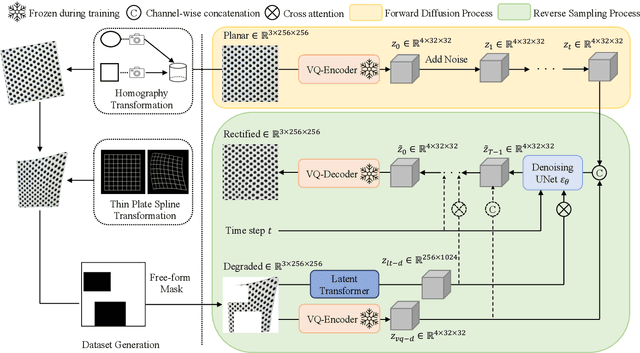
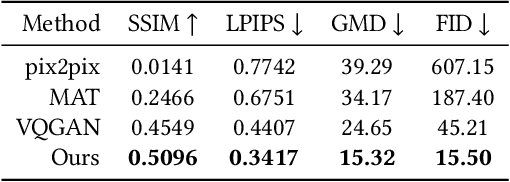
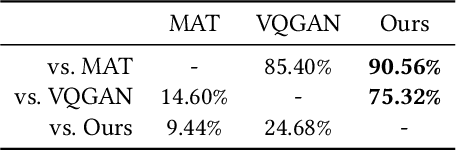
Abstract:We present a novel framework for rectifying occlusions and distortions in degraded texture samples from natural images. Traditional texture synthesis approaches focus on generating textures from pristine samples, which necessitate meticulous preparation by humans and are often unattainable in most natural images. These challenges stem from the frequent occlusions and distortions of texture samples in natural images due to obstructions and variations in object surface geometry. To address these issues, we propose a framework that synthesizes holistic textures from degraded samples in natural images, extending the applicability of exemplar-based texture synthesis techniques. Our framework utilizes a conditional Latent Diffusion Model (LDM) with a novel occlusion-aware latent transformer. This latent transformer not only effectively encodes texture features from partially-observed samples necessary for the generation process of the LDM, but also explicitly captures long-range dependencies in samples with large occlusions. To train our model, we introduce a method for generating synthetic data by applying geometric transformations and free-form mask generation to clean textures. Experimental results demonstrate that our framework significantly outperforms existing methods both quantitatively and quantitatively. Furthermore, we conduct comprehensive ablation studies to validate the different components of our proposed framework. Results are corroborated by a perceptual user study which highlights the efficiency of our proposed approach.
Controllable Multi-domain Semantic Artwork Synthesis
Aug 19, 2023Abstract:We present a novel framework for multi-domain synthesis of artwork from semantic layouts. One of the main limitations of this challenging task is the lack of publicly available segmentation datasets for art synthesis. To address this problem, we propose a dataset, which we call ArtSem, that contains 40,000 images of artwork from 4 different domains with their corresponding semantic label maps. We generate the dataset by first extracting semantic maps from landscape photography and then propose a conditional Generative Adversarial Network (GAN)-based approach to generate high-quality artwork from the semantic maps without necessitating paired training data. Furthermore, we propose an artwork synthesis model that uses domain-dependent variational encoders for high-quality multi-domain synthesis. The model is improved and complemented with a simple but effective normalization method, based on normalizing both the semantic and style jointly, which we call Spatially STyle-Adaptive Normalization (SSTAN). In contrast to previous methods that only take semantic layout as input, our model is able to learn a joint representation of both style and semantic information, which leads to better generation quality for synthesizing artistic images. Results indicate that our model learns to separate the domains in the latent space, and thus, by identifying the hyperplanes that separate the different domains, we can also perform fine-grained control of the synthesized artwork. By combining our proposed dataset and approach, we are able to generate user-controllable artwork that is of higher quality than existing
P2Net: A Post-Processing Network for Refining Semantic Segmentation of LiDAR Point Cloud based on Consistency of Consecutive Frames
Dec 01, 2022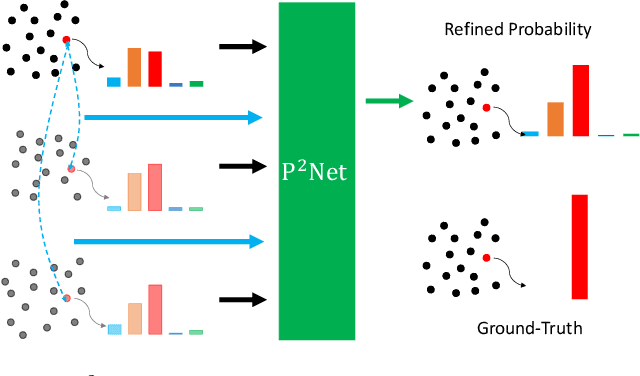
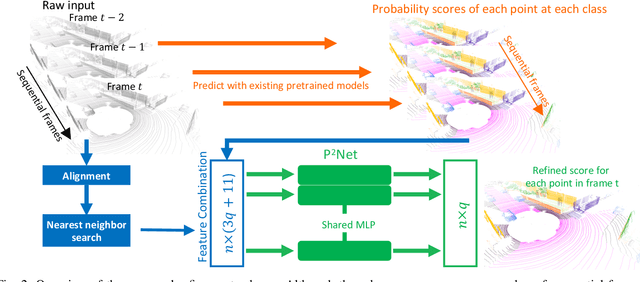
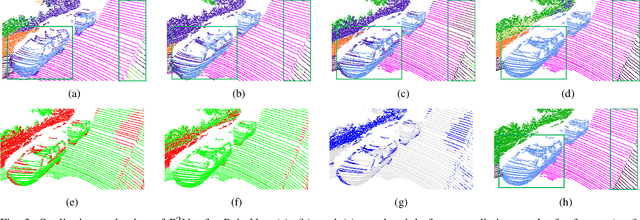
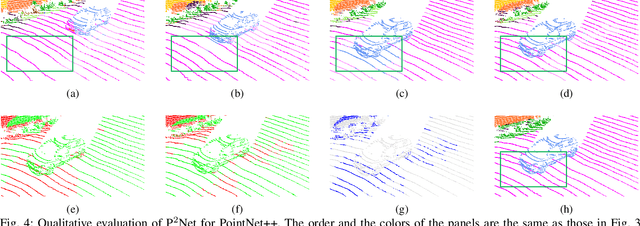
Abstract:We present a lightweight post-processing method to refine the semantic segmentation results of point cloud sequences. Most existing methods usually segment frame by frame and encounter the inherent ambiguity of the problem: based on a measurement in a single frame, labels are sometimes difficult to predict even for humans. To remedy this problem, we propose to explicitly train a network to refine these results predicted by an existing segmentation method. The network, which we call the P2Net, learns the consistency constraints between coincident points from consecutive frames after registration. We evaluate the proposed post-processing method both qualitatively and quantitatively on the SemanticKITTI dataset that consists of real outdoor scenes. The effectiveness of the proposed method is validated by comparing the results predicted by two representative networks with and without the refinement by the post-processing network. Specifically, qualitative visualization validates the key idea that labels of the points that are difficult to predict can be corrected with P2Net. Quantitatively, overall mIoU is improved from 10.5% to 11.7% for PointNet [1] and from 10.8% to 15.9% for PointNet++ [2].
Automatic Segmentation, Localization, and Identification of Vertebrae in 3D CT Images Using Cascaded Convolutional Neural Networks
Sep 29, 2020
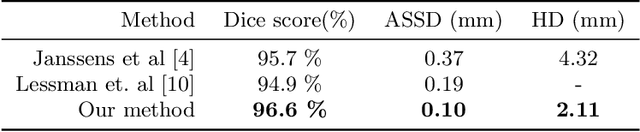
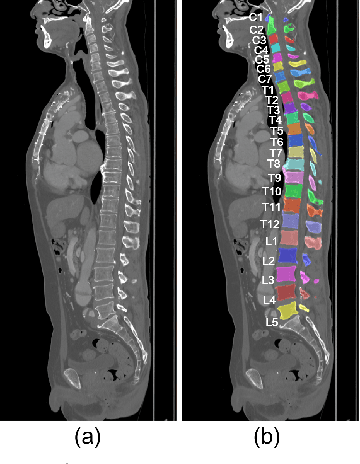
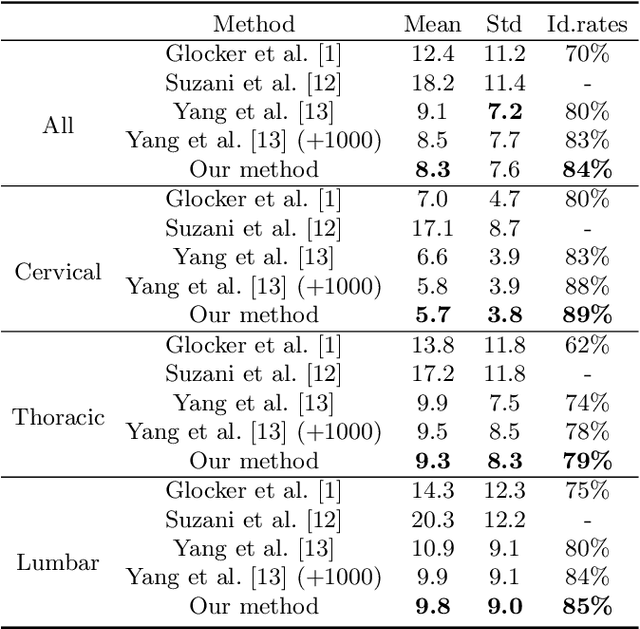
Abstract:This paper presents a method for automatic segmentation, localization, and identification of vertebrae in arbitrary 3D CT images. Many previous works do not perform the three tasks simultaneously even though requiring a priori knowledge of which part of the anatomy is visible in the 3D CT images. Our method tackles all these tasks in a single multi-stage framework without any assumptions. In the first stage, we train a 3D Fully Convolutional Networks to find the bounding boxes of the cervical, thoracic, and lumbar vertebrae. In the second stage, we train an iterative 3D Fully Convolutional Networks to segment individual vertebrae in the bounding box. The input to the second networks have an auxiliary channel in addition to the 3D CT images. Given the segmented vertebra regions in the auxiliary channel, the networks output the next vertebra. The proposed method is evaluated in terms of segmentation, localization, and identification accuracy with two public datasets of 15 3D CT images from the MICCAI CSI 2014 workshop challenge and 302 3D CT images with various pathologies introduced in [1]. Our method achieved a mean Dice score of 96%, a mean localization error of 8.3 mm, and a mean identification rate of 84%. In summary, our method achieved better performance than all existing works in all the three metrics.
DeepRemaster: Temporal Source-Reference Attention Networks for Comprehensive Video Enhancement
Sep 18, 2020
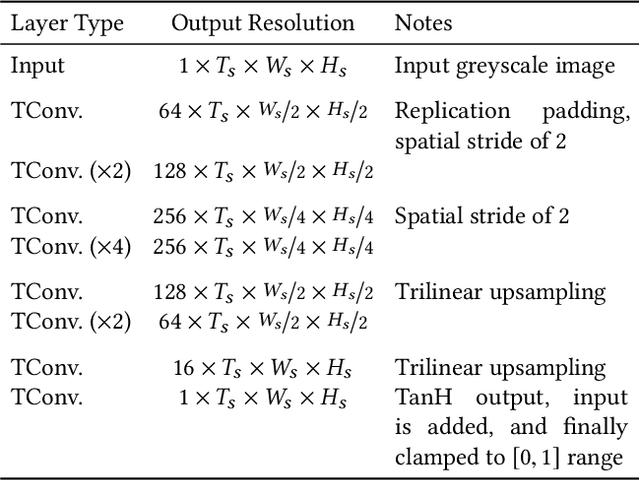

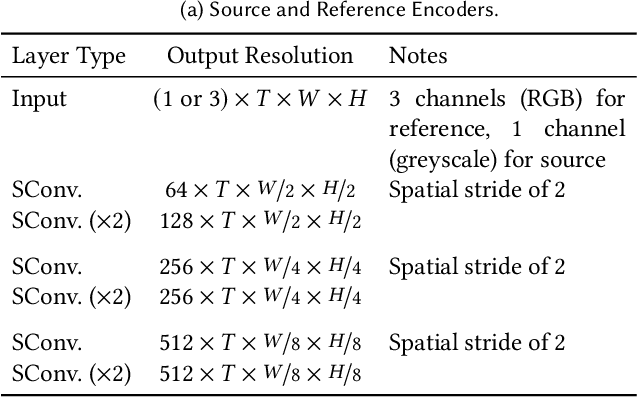
Abstract:The remastering of vintage film comprises of a diversity of sub-tasks including super-resolution, noise removal, and contrast enhancement which aim to restore the deteriorated film medium to its original state. Additionally, due to the technical limitations of the time, most vintage film is either recorded in black and white, or has low quality colors, for which colorization becomes necessary. In this work, we propose a single framework to tackle the entire remastering task semi-interactively. Our work is based on temporal convolutional neural networks with attention mechanisms trained on videos with data-driven deterioration simulation. Our proposed source-reference attention allows the model to handle an arbitrary number of reference color images to colorize long videos without the need for segmentation while maintaining temporal consistency. Quantitative analysis shows that our framework outperforms existing approaches, and that, in contrast to existing approaches, the performance of our framework increases with longer videos and more reference color images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge