Noriyuki Tomiyama
Visual Grounding of Whole Radiology Reports for 3D CT Images
Dec 08, 2023Abstract:Building a large-scale training dataset is an essential problem in the development of medical image recognition systems. Visual grounding techniques, which automatically associate objects in images with corresponding descriptions, can facilitate labeling of large number of images. However, visual grounding of radiology reports for CT images remains challenging, because so many kinds of anomalies are detectable via CT imaging, and resulting report descriptions are long and complex. In this paper, we present the first visual grounding framework designed for CT image and report pairs covering various body parts and diverse anomaly types. Our framework combines two components of 1) anatomical segmentation of images, and 2) report structuring. The anatomical segmentation provides multiple organ masks of given CT images, and helps the grounding model recognize detailed anatomies. The report structuring helps to accurately extract information regarding the presence, location, and type of each anomaly described in corresponding reports. Given the two additional image/report features, the grounding model can achieve better localization. In the verification process, we constructed a large-scale dataset with region-description correspondence annotations for 10,410 studies of 7,321 unique patients. We evaluated our framework using grounding accuracy, the percentage of correctly localized anomalies, as a metric and demonstrated that the combination of the anatomical segmentation and the report structuring improves the performance with a large margin over the baseline model (66.0% vs 77.8%). Comparison with the prior techniques also showed higher performance of our method.
* 14 pages, 7 figures. Accepted at MICCAI 2023
Anatomy-aware Self-supervised Learning for Anomaly Detection in Chest Radiographs
May 09, 2022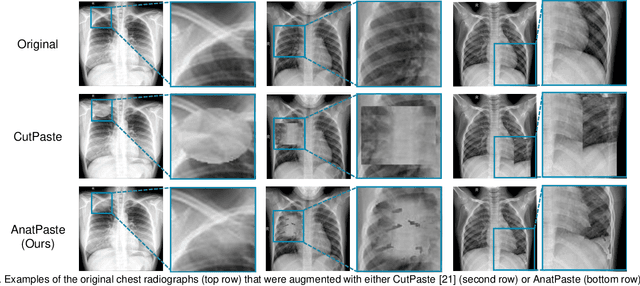
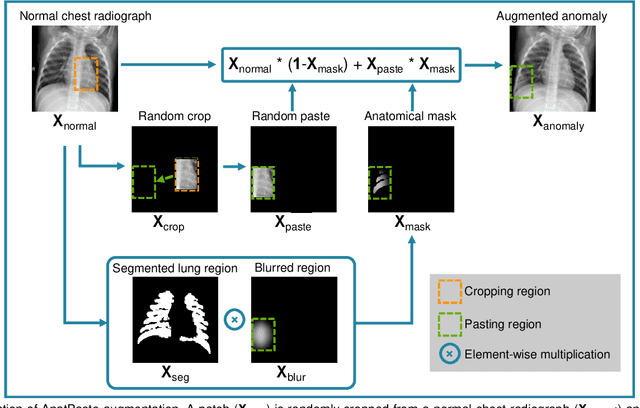
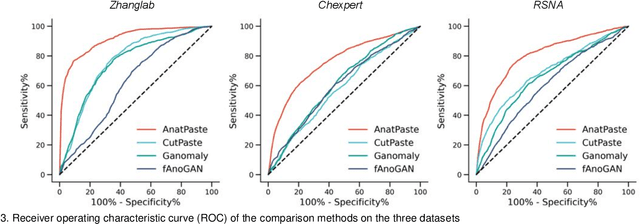
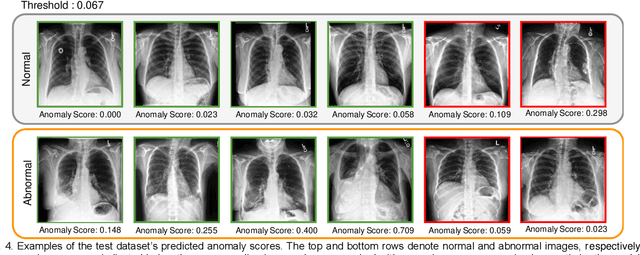
Abstract:Large numbers of labeled medical images are essential for the accurate detection of anomalies, but manual annotation is labor-intensive and time-consuming. Self-supervised learning (SSL) is a training method to learn data-specific features without manual annotation. Several SSL-based models have been employed in medical image anomaly detection. These SSL methods effectively learn representations in several field-specific images, such as natural and industrial product images. However, owing to the requirement of medical expertise, typical SSL-based models are inefficient in medical image anomaly detection. We present an SSL-based model that enables anatomical structure-based unsupervised anomaly detection (UAD). The model employs the anatomy-aware pasting (AnatPaste) augmentation tool. AnatPaste employs a threshold-based lung segmentation pretext task to create anomalies in normal chest radiographs, which are used for model pretraining. These anomalies are similar to real anomalies and help the model recognize them. We evaluate our model on three opensource chest radiograph datasets. Our model exhibit area under curves (AUC) of 92.1%, 78.7%, and 81.9%, which are the highest among existing UAD models. This is the first SSL model to employ anatomical information as a pretext task. AnatPaste can be applied in various deep learning models and downstream tasks. It can be employed for other modalities by fixing appropriate segmentation. Our code is publicly available at: https://github.com/jun-sato/AnatPaste.
Weak Supervision in Convolutional Neural Network for Semantic Segmentation of Diffuse Lung Diseases Using Partially Annotated Dataset
Mar 26, 2020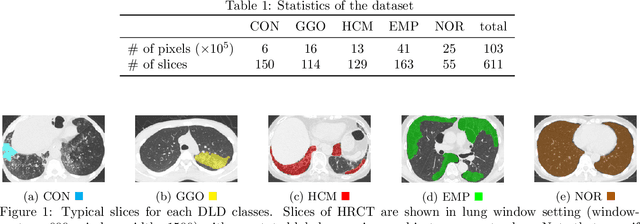
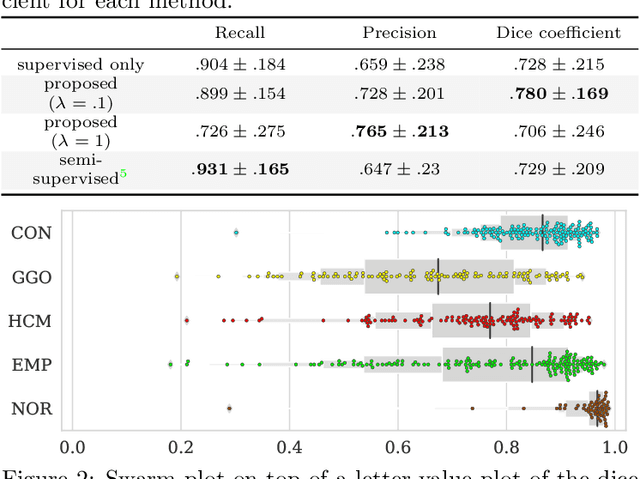
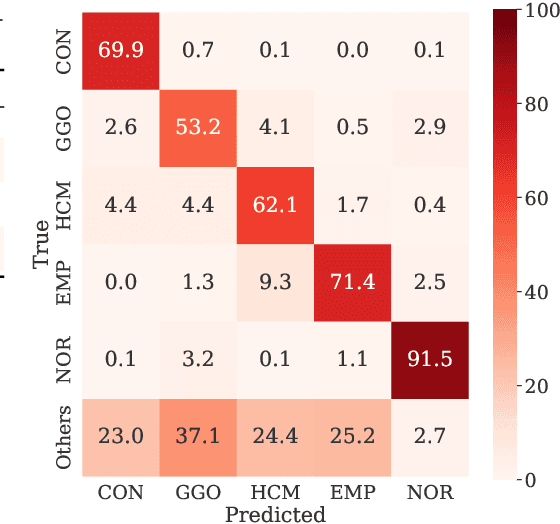
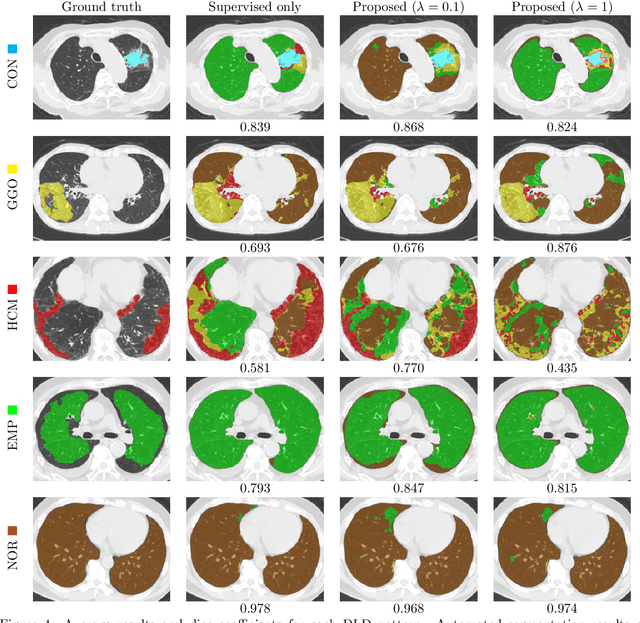
Abstract:Computer-aided diagnosis system for diffuse lung diseases (DLDs) is necessary for the objective assessment of the lung diseases. In this paper, we develop semantic segmentation model for 5 kinds of DLDs. DLDs considered in this work are consolidation, ground glass opacity, honeycombing, emphysema, and normal. Convolutional neural network (CNN) is one of the most promising technique for semantic segmentation among machine learning algorithms. While creating annotated dataset for semantic segmentation is laborious and time consuming, creating partially annotated dataset, in which only one chosen class is annotated for each image, is easier since annotators only need to focus on one class at a time during the annotation task. In this paper, we propose a new weak supervision technique that effectively utilizes partially annotated dataset. The experiments using partially annotated dataset composed 372 CT images demonstrated that our proposed technique significantly improved segmentation accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge