Keigo Nakamura
Visual Grounding of Whole Radiology Reports for 3D CT Images
Dec 08, 2023Abstract:Building a large-scale training dataset is an essential problem in the development of medical image recognition systems. Visual grounding techniques, which automatically associate objects in images with corresponding descriptions, can facilitate labeling of large number of images. However, visual grounding of radiology reports for CT images remains challenging, because so many kinds of anomalies are detectable via CT imaging, and resulting report descriptions are long and complex. In this paper, we present the first visual grounding framework designed for CT image and report pairs covering various body parts and diverse anomaly types. Our framework combines two components of 1) anatomical segmentation of images, and 2) report structuring. The anatomical segmentation provides multiple organ masks of given CT images, and helps the grounding model recognize detailed anatomies. The report structuring helps to accurately extract information regarding the presence, location, and type of each anomaly described in corresponding reports. Given the two additional image/report features, the grounding model can achieve better localization. In the verification process, we constructed a large-scale dataset with region-description correspondence annotations for 10,410 studies of 7,321 unique patients. We evaluated our framework using grounding accuracy, the percentage of correctly localized anomalies, as a metric and demonstrated that the combination of the anatomical segmentation and the report structuring improves the performance with a large margin over the baseline model (66.0% vs 77.8%). Comparison with the prior techniques also showed higher performance of our method.
* 14 pages, 7 figures. Accepted at MICCAI 2023
Segmentation of Kidney Tumors on Non-Contrast CT Images using Protuberance Detection Network
Dec 08, 2023Abstract:Many renal cancers are incidentally found on non-contrast CT (NCCT) images. On contrast-enhanced CT (CECT) images, most kidney tumors, especially renal cancers, have different intensity values compared to normal tissues. However, on NCCT images, some tumors called isodensity tumors, have similar intensity values to the surrounding normal tissues, and can only be detected through a change in organ shape. Several deep learning methods which segment kidney tumors from CECT images have been proposed and showed promising results. However, these methods fail to capture such changes in organ shape on NCCT images. In this paper, we present a novel framework, which can explicitly capture protruded regions in kidneys to enable a better segmentation of kidney tumors. We created a synthetic mask dataset that simulates a protuberance, and trained a segmentation network to separate the protruded regions from the normal kidney regions. To achieve the segmentation of whole tumors, our framework consists of three networks. The first network is a conventional semantic segmentation network which extracts a kidney region mask and an initial tumor region mask. The second network, which we name protuberance detection network, identifies the protruded regions from the kidney region mask. Given the initial tumor region mask and the protruded region mask, the last network fuses them and predicts the final kidney tumor mask accurately. The proposed method was evaluated on a publicly available KiTS19 dataset, which contains 108 NCCT images, and showed that our method achieved a higher dice score of 0.615 (+0.097) and sensitivity of 0.721 (+0.103) compared to 3D-UNet. To the best of our knowledge, this is the first deep learning method that is specifically designed for kidney tumor segmentation on NCCT images.
* Accepted in MICCAI 2023
C-SENN: Contrastive Self-Explaining Neural Network
Jun 27, 2022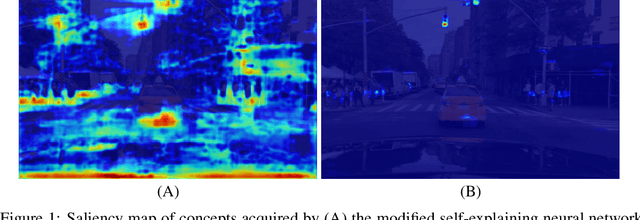



Abstract:In this study, we use a self-explaining neural network (SENN), which learns unsupervised concepts, to acquire concepts that are easy for people to understand automatically. In concept learning, the hidden layer retains verbalizable features relevant to the output, which is crucial when adapting to real-world environments where explanations are required. However, it is known that the interpretability of concepts output by SENN is reduced in general settings, such as autonomous driving scenarios. Thus, this study combines contrastive learning with concept learning to improve the readability of concepts and the accuracy of tasks. We call this model Contrastive Self-Explaining Neural Network (C-SENN).
Concept Bottleneck Model with Additional Unsupervised Concepts
Feb 03, 2022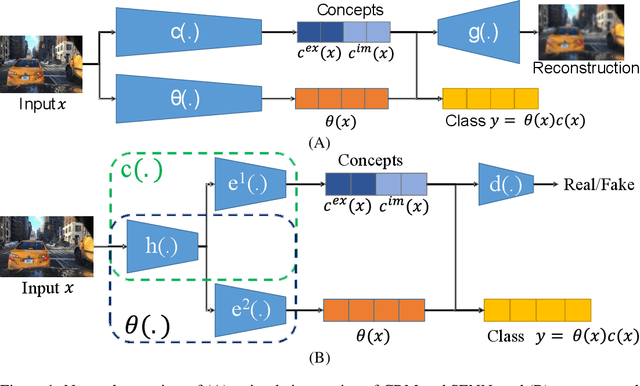
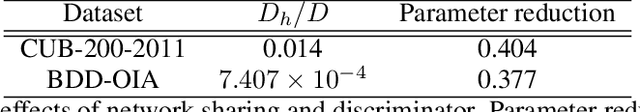
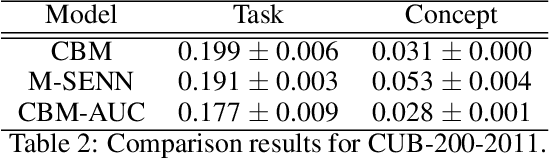
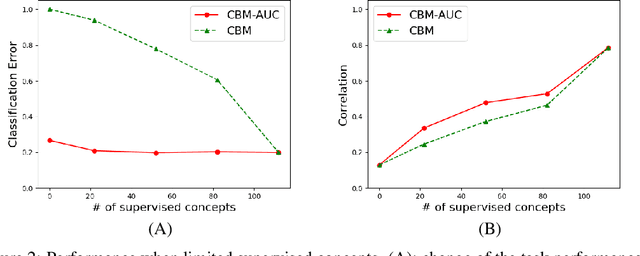
Abstract:With the increasing demands for accountability, interpretability is becoming an essential capability for real-world AI applications. However, most methods utilize post-hoc approaches rather than training the interpretable model. In this article, we propose a novel interpretable model based on the concept bottleneck model (CBM). CBM uses concept labels to train an intermediate layer as the additional visible layer. However, because the number of concept labels restricts the dimension of this layer, it is difficult to obtain high accuracy with a small number of labels. To address this issue, we integrate supervised concepts with unsupervised ones trained with self-explaining neural networks (SENNs). By seamlessly training these two types of concepts while reducing the amount of computation, we can obtain both supervised and unsupervised concepts simultaneously, even for large-sized images. We refer to the proposed model as the concept bottleneck model with additional unsupervised concepts (CBM-AUC). We experimentally confirmed that the proposed model outperformed CBM and SENN. We also visualized the saliency map of each concept and confirmed that it was consistent with the semantic meanings.
Automatic Segmentation, Localization, and Identification of Vertebrae in 3D CT Images Using Cascaded Convolutional Neural Networks
Sep 29, 2020
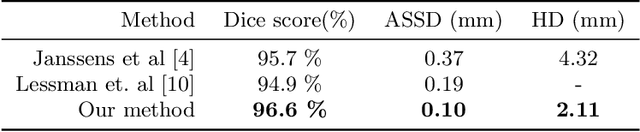
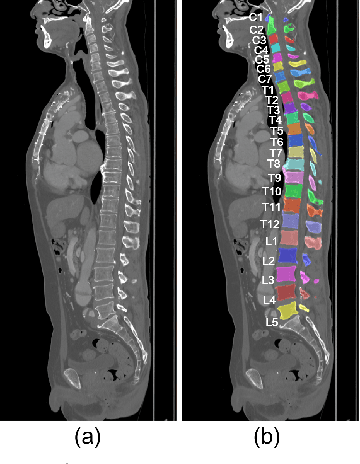
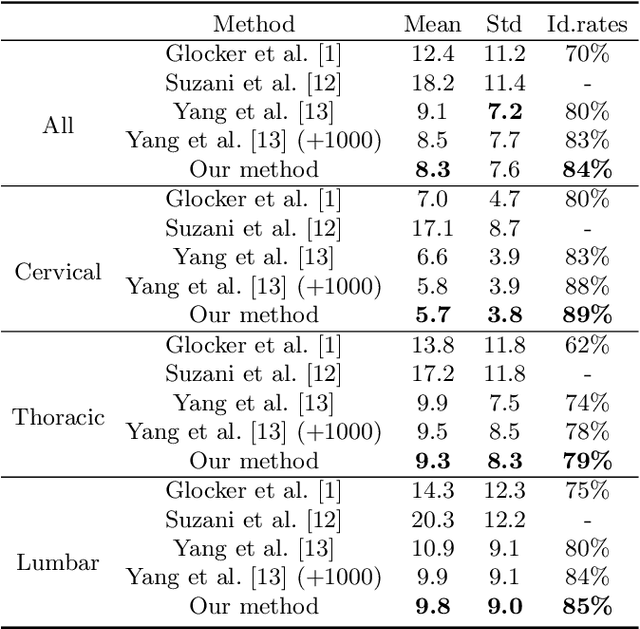
Abstract:This paper presents a method for automatic segmentation, localization, and identification of vertebrae in arbitrary 3D CT images. Many previous works do not perform the three tasks simultaneously even though requiring a priori knowledge of which part of the anatomy is visible in the 3D CT images. Our method tackles all these tasks in a single multi-stage framework without any assumptions. In the first stage, we train a 3D Fully Convolutional Networks to find the bounding boxes of the cervical, thoracic, and lumbar vertebrae. In the second stage, we train an iterative 3D Fully Convolutional Networks to segment individual vertebrae in the bounding box. The input to the second networks have an auxiliary channel in addition to the 3D CT images. Given the segmented vertebra regions in the auxiliary channel, the networks output the next vertebra. The proposed method is evaluated in terms of segmentation, localization, and identification accuracy with two public datasets of 15 3D CT images from the MICCAI CSI 2014 workshop challenge and 302 3D CT images with various pathologies introduced in [1]. Our method achieved a mean Dice score of 96%, a mean localization error of 8.3 mm, and a mean identification rate of 84%. In summary, our method achieved better performance than all existing works in all the three metrics.
Deep learning-based topological optimization for representing a user-specified design area
Apr 19, 2020

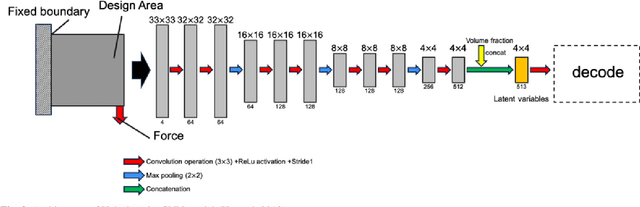

Abstract:Presently, topology optimization requires multiple iterations to create an optimized structure for given conditions. Among the conditions for topology optimization,the design area is one of the most important for structural design. In this study, we propose a new deep learning model to generate an optimized structure for a given design domain and other boundary conditions without iteration. For this purpose, we used open-source topology optimization MATLAB code to generate a pair of optimized structures under various design conditions. The resolution of the optimized structure is 32 * 32 pixels, and the design conditions are design area, volume fraction, distribution of external forces, and load value. Our deep learning model is primarily composed of a convolutional neural network (CNN)-based encoder and decoder, trained with datasets generated with MATLAB code. In the encoder, we use batch normalization (BN) to increase the stability of the CNN model. In the decoder, we use SPADE (spatially adaptive denormalization) to reinforce the design area information. Comparing the performance of our proposed model with a CNN model that does not use BN and SPADE, values for mean absolute error (MAE), mean compliance error, and volume error with the optimized topology structure generated in MAT-LAB code were smaller, and the proposed model was able to represent the design area more precisely. The proposed method generates near-optimal structures reflecting the design area in less computational time, compared with the open-source topology optimization MATLAB code.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge