Yoshiro Kitamura
Data-Dependent Higher-Order Clique Selection for Artery-Vein Segmentation by Energy Minimization
Dec 13, 2023Abstract:We propose a novel segmentation method based on energy minimization of higher-order potentials. We introduce higher-order terms into the energy to incorporate prior knowledge on the shape of the segments. The terms encourage certain sets of pixels to be entirely in one segment or the other. The sets can for instance be smooth curves in order to help delineate pulmonary vessels, which are known to run in almost straight lines. The higher-order terms can be converted to submodular first-order terms by adding auxiliary variables, which can then be globally minimized using graph cuts. We also determine the weight of these terms, or the degree of the aforementioned encouragement, in a principled way by learning from training data with the ground truth. We demonstrate the effectiveness of the method in a real-world application in fully-automatic pulmonary artery-vein segmentation in CT images.
Visual Grounding of Whole Radiology Reports for 3D CT Images
Dec 08, 2023Abstract:Building a large-scale training dataset is an essential problem in the development of medical image recognition systems. Visual grounding techniques, which automatically associate objects in images with corresponding descriptions, can facilitate labeling of large number of images. However, visual grounding of radiology reports for CT images remains challenging, because so many kinds of anomalies are detectable via CT imaging, and resulting report descriptions are long and complex. In this paper, we present the first visual grounding framework designed for CT image and report pairs covering various body parts and diverse anomaly types. Our framework combines two components of 1) anatomical segmentation of images, and 2) report structuring. The anatomical segmentation provides multiple organ masks of given CT images, and helps the grounding model recognize detailed anatomies. The report structuring helps to accurately extract information regarding the presence, location, and type of each anomaly described in corresponding reports. Given the two additional image/report features, the grounding model can achieve better localization. In the verification process, we constructed a large-scale dataset with region-description correspondence annotations for 10,410 studies of 7,321 unique patients. We evaluated our framework using grounding accuracy, the percentage of correctly localized anomalies, as a metric and demonstrated that the combination of the anatomical segmentation and the report structuring improves the performance with a large margin over the baseline model (66.0% vs 77.8%). Comparison with the prior techniques also showed higher performance of our method.
* 14 pages, 7 figures. Accepted at MICCAI 2023
Image Synthesis-based Late Stage Cancer Augmentation and Semi-Supervised Segmentation for MRI Rectal Cancer Staging
Dec 08, 2023Abstract:Rectal cancer is one of the most common diseases and a major cause of mortality. For deciding rectal cancer treatment plans, T-staging is important. However, evaluating the index from preoperative MRI images requires high radiologists' skill and experience. Therefore, the aim of this study is to segment the mesorectum, rectum, and rectal cancer region so that the system can predict T-stage from segmentation results. Generally, shortage of large and diverse dataset and high quality annotation are known to be the bottlenecks in computer aided diagnostics development. Regarding rectal cancer, advanced cancer images are very rare, and per-pixel annotation requires high radiologists' skill and time. Therefore, it is not feasible to collect comprehensive disease patterns in a training dataset. To tackle this, we propose two kinds of approaches of image synthesis-based late stage cancer augmentation and semi-supervised learning which is designed for T-stage prediction. In the image synthesis data augmentation approach, we generated advanced cancer images from labels. The real cancer labels were deformed to resemble advanced cancer labels by artificial cancer progress simulation. Next, we introduce a T-staging loss which enables us to train segmentation models from per-image T-stage labels. The loss works to keep inclusion/invasion relationships between rectum and cancer region consistent to the ground truth T-stage. The verification tests show that the proposed method obtains the best sensitivity (0.76) and specificity (0.80) in distinguishing between over T3 stage and underT2. In the ablation studies, our semi-supervised learning approach with the T-staging loss improved specificity by 0.13. Adding the image synthesis-based data augmentation improved the DICE score of invasion cancer area by 0.08 from baseline.
Segmentation of Kidney Tumors on Non-Contrast CT Images using Protuberance Detection Network
Dec 08, 2023Abstract:Many renal cancers are incidentally found on non-contrast CT (NCCT) images. On contrast-enhanced CT (CECT) images, most kidney tumors, especially renal cancers, have different intensity values compared to normal tissues. However, on NCCT images, some tumors called isodensity tumors, have similar intensity values to the surrounding normal tissues, and can only be detected through a change in organ shape. Several deep learning methods which segment kidney tumors from CECT images have been proposed and showed promising results. However, these methods fail to capture such changes in organ shape on NCCT images. In this paper, we present a novel framework, which can explicitly capture protruded regions in kidneys to enable a better segmentation of kidney tumors. We created a synthetic mask dataset that simulates a protuberance, and trained a segmentation network to separate the protruded regions from the normal kidney regions. To achieve the segmentation of whole tumors, our framework consists of three networks. The first network is a conventional semantic segmentation network which extracts a kidney region mask and an initial tumor region mask. The second network, which we name protuberance detection network, identifies the protruded regions from the kidney region mask. Given the initial tumor region mask and the protruded region mask, the last network fuses them and predicts the final kidney tumor mask accurately. The proposed method was evaluated on a publicly available KiTS19 dataset, which contains 108 NCCT images, and showed that our method achieved a higher dice score of 0.615 (+0.097) and sensitivity of 0.721 (+0.103) compared to 3D-UNet. To the best of our knowledge, this is the first deep learning method that is specifically designed for kidney tumor segmentation on NCCT images.
* Accepted in MICCAI 2023
Automatic Segmentation, Localization, and Identification of Vertebrae in 3D CT Images Using Cascaded Convolutional Neural Networks
Sep 29, 2020
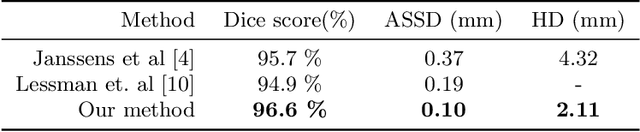
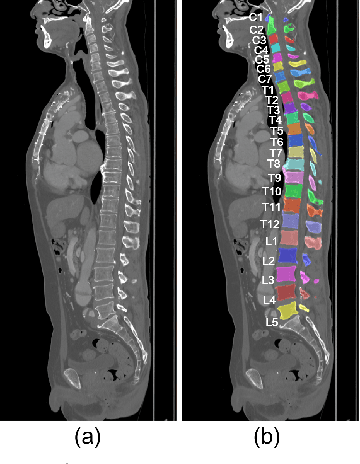
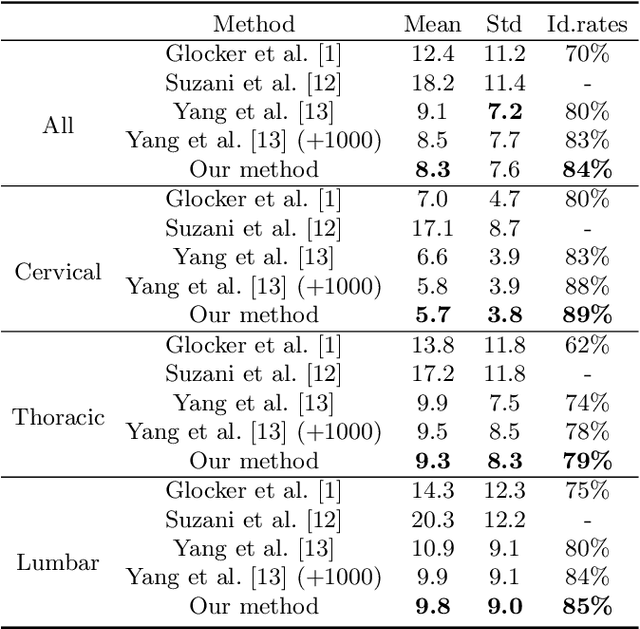
Abstract:This paper presents a method for automatic segmentation, localization, and identification of vertebrae in arbitrary 3D CT images. Many previous works do not perform the three tasks simultaneously even though requiring a priori knowledge of which part of the anatomy is visible in the 3D CT images. Our method tackles all these tasks in a single multi-stage framework without any assumptions. In the first stage, we train a 3D Fully Convolutional Networks to find the bounding boxes of the cervical, thoracic, and lumbar vertebrae. In the second stage, we train an iterative 3D Fully Convolutional Networks to segment individual vertebrae in the bounding box. The input to the second networks have an auxiliary channel in addition to the 3D CT images. Given the segmented vertebra regions in the auxiliary channel, the networks output the next vertebra. The proposed method is evaluated in terms of segmentation, localization, and identification accuracy with two public datasets of 15 3D CT images from the MICCAI CSI 2014 workshop challenge and 302 3D CT images with various pathologies introduced in [1]. Our method achieved a mean Dice score of 96%, a mean localization error of 8.3 mm, and a mean identification rate of 84%. In summary, our method achieved better performance than all existing works in all the three metrics.
TopNet: Topology Preserving Metric Learning for Vessel Tree Reconstruction and Labelling
Sep 18, 2020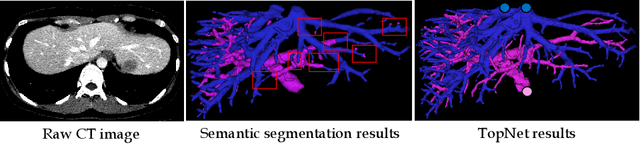

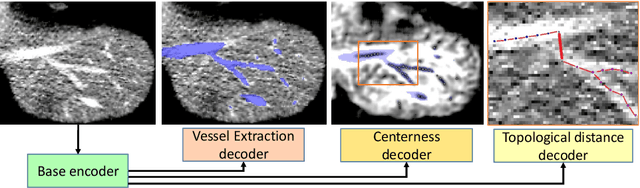

Abstract:Reconstructing Portal Vein and Hepatic Vein trees from contrast enhanced abdominal CT scans is a prerequisite for preoperative liver surgery simulation. Existing deep learning based methods treat vascular tree reconstruction as a semantic segmentation problem. However, vessels such as hepatic and portal vein look very similar locally and need to be traced to their source for robust label assignment. Therefore, semantic segmentation by looking at local 3D patch results in noisy misclassifications. To tackle this, we propose a novel multi-task deep learning architecture for vessel tree reconstruction. The network architecture simultaneously solves the task of detecting voxels on vascular centerlines (i.e. nodes) and estimates connectivity between center-voxels (edges) in the tree structure to be reconstructed. Further, we propose a novel connectivity metric which considers both inter-class distance and intra-class topological distance between center-voxel pairs. Vascular trees are reconstructed starting from the vessel source using the learned connectivity metric using the shortest path tree algorithm. A thorough evaluation on public IRCAD dataset shows that the proposed method considerably outperforms existing semantic segmentation based methods. To the best of our knowledge, this is the first deep learning based approach which learns multi-label tree structure connectivity from images.
Virtual Thin Slice: 3D Conditional GAN-based Super-resolution for CT Slice Interval
Sep 02, 2019
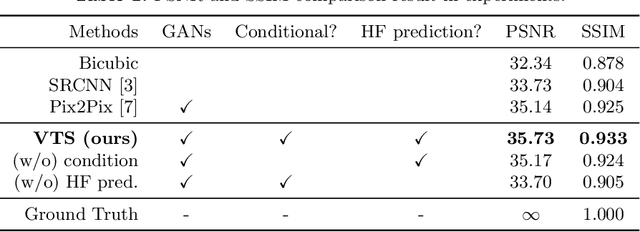
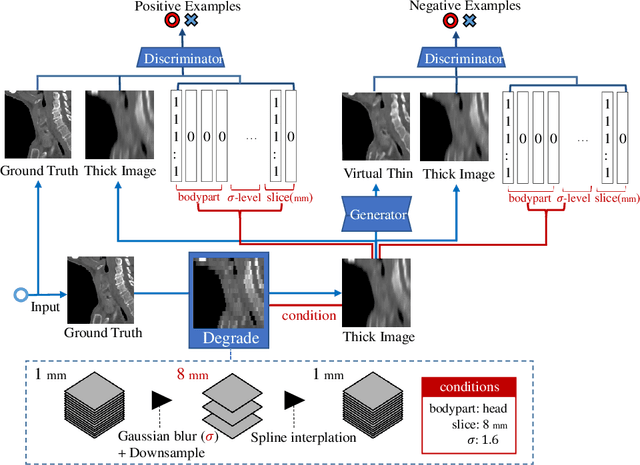
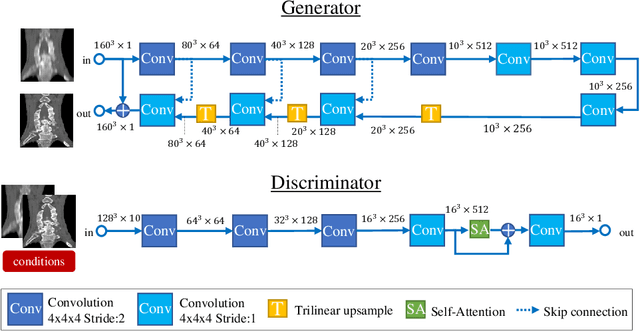
Abstract:Many CT slice images are stored with large slice intervals to reduce storage size in clinical practice. This leads to low resolution perpendicular to the slice images (i.e., z-axis), which is insufficient for 3D visualization or image analysis. In this paper, we present a novel architecture based on conditional Generative Adversarial Networks (cGANs) with the goal of generating high resolution images of main body parts including head, chest, abdomen and legs. However, GANs are known to have a difficulty with generating a diversity of patterns due to a phenomena known as mode collapse. To overcome the lack of generated pattern variety, we propose to condition the discriminator on the different body parts. Furthermore, our generator networks are extended to be three dimensional fully convolutional neural networks, allowing for the generation of high resolution images from arbitrary fields of view. In our verification tests, we show that the proposed method obtains the best scores by PSNR/SSIM metrics and Visual Turing Test, allowing for accurate reproduction of the principle anatomy in high resolution. We expect that the proposed method contribute to effective utilization of the existing vast amounts of thick CT images stored in hospitals.
Synthesizing Diverse Lung Nodules Wherever Massively: 3D Multi-Conditional GAN-based CT Image Augmentation for Object Detection
Jun 12, 2019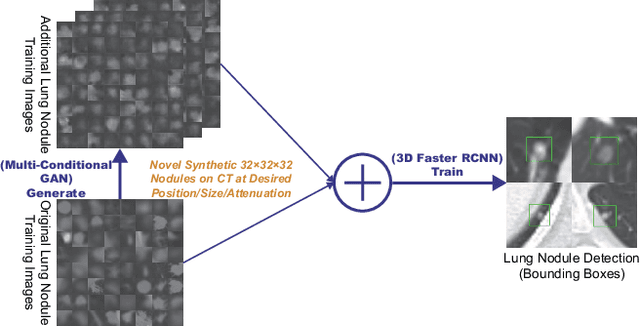
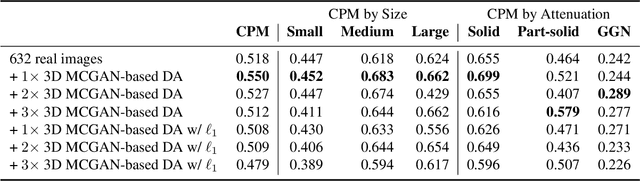
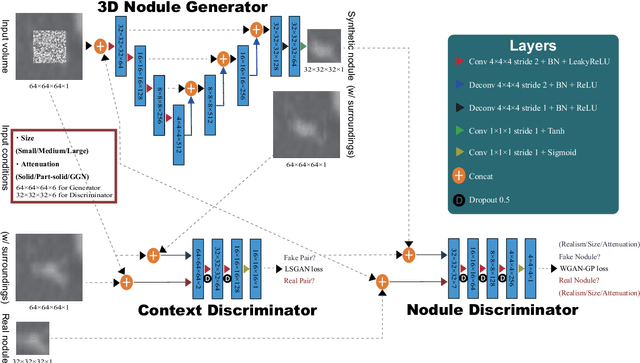
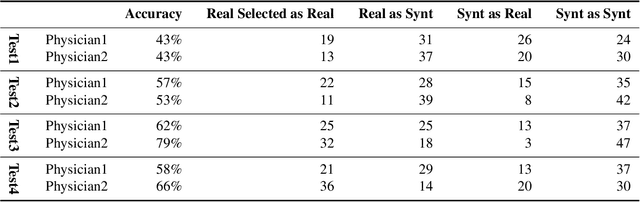
Abstract:Accurate computer-assisted diagnosis, relying on large-scale annotated pathological images, can alleviate the risk of overlooking the diagnosis. Unfortunately, in medical imaging, most available datasets are small/fragmented. To tackle this, as a Data Augmentation (DA) method, 3D conditional Generative Adversarial Networks (GANs) can synthesize desired realistic/diverse 3D images as additional training data. However, no 3D conditional GAN-based DA approach exists for general bounding box-based 3D object detection, while it can locate disease areas with physicians' minimum annotation cost, unlike rigorous 3D segmentation. Moreover, since lesions vary in position/size/attenuation, further GAN-based DA performance requires multiple conditions. Therefore, we propose 3D Multi-Conditional GAN (MCGAN) to generate realistic/diverse 32 x 32 x 32 nodules placed naturally on lung Computed Tomography images to boost sensitivity in 3D object detection. Our MCGAN adopts two discriminators for conditioning: the context discriminator learns to classify real vs synthetic nodule/surrounding pairs with noise box-centered surroundings; the nodule discriminator attempts to classify real vs synthetic nodules with size/attenuation conditions. The results show that 3D Convolutional Neural Network-based detection can achieve higher sensitivity under any nodule size/attenuation at fixed False Positive rates and overcome the medical data paucity with the MCGAN-generated realistic nodules---even expert physicians fail to distinguish them from the real ones in Visual Turing Test.
Computation of Total Kidney Volume from CT images in Autosomal Dominant Polycystic Kidney Disease using Multi-Task 3D Convolutional Neural Networks
Sep 07, 2018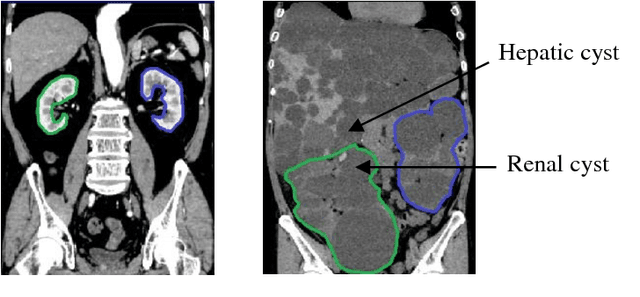
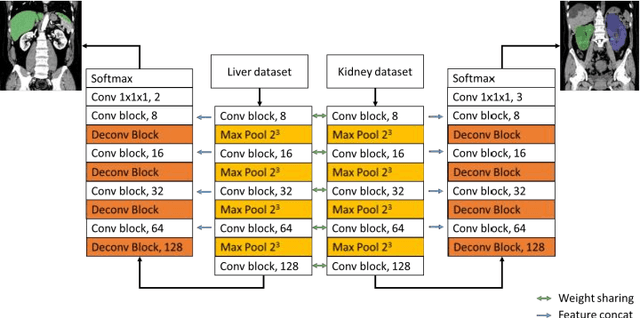

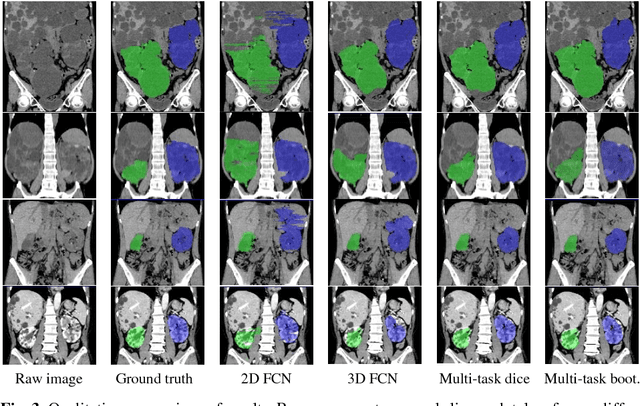
Abstract:Autosomal Dominant Polycystic Kidney Disease (ADPKD) characterized by progressive growth of renal cysts is the most prevalent and potentially lethal monogenic renal disease, affecting one in every 500-100 people. Total Kidney Volume (TKV) and its growth computed from Computed Tomography images has been accepted as an essential prognostic marker for renal function loss. Due to large variation in shape and size of kidney in ADPKD, existing methods to compute TKV (i.e. to segment ADKP) including those based on 2D convolutional neural networks are not accurate enough to be directly useful in clinical practice. In this work, we propose multi-task 3D Convolutional Neural Networks to segment ADPK and achieve a mean DICE score of 0.95 and mean absolute percentage TKV error of 3.86. Additionally, to solve the challenge of class imbalance, we propose to simply bootstrap cross entropy loss and compare results with recently prevalent dice loss in medical image segmentation community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge