Samaneh Kouchaki
Interpreting Differentiable Latent States for Healthcare Time-series Data
Nov 29, 2023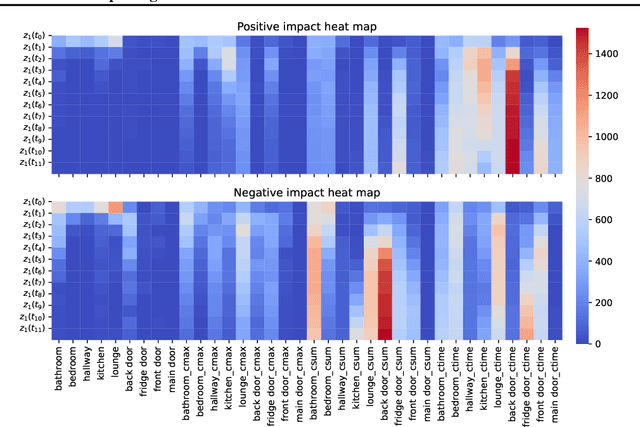

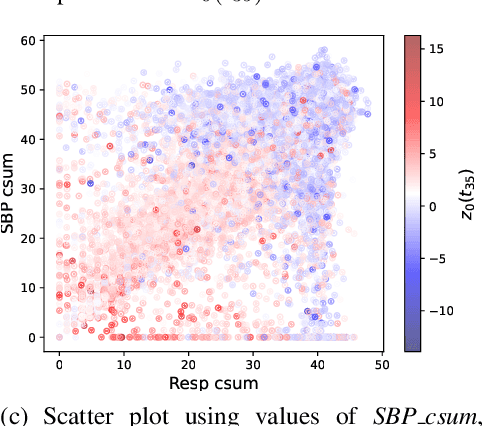
Abstract:Machine learning enables extracting clinical insights from large temporal datasets. The applications of such machine learning models include identifying disease patterns and predicting patient outcomes. However, limited interpretability poses challenges for deploying advanced machine learning in digital healthcare. Understanding the meaning of latent states is crucial for interpreting machine learning models, assuming they capture underlying patterns. In this paper, we present a concise algorithm that allows for i) interpreting latent states using highly related input features; ii) interpreting predictions using subsets of input features via latent states; and iii) interpreting changes in latent states over time. The proposed algorithm is feasible for any model that is differentiable. We demonstrate that this approach enables the identification of a daytime behavioral pattern for predicting nocturnal behavior in a real-world healthcare dataset.
Lightweight Transformers for Clinical Natural Language Processing
Feb 09, 2023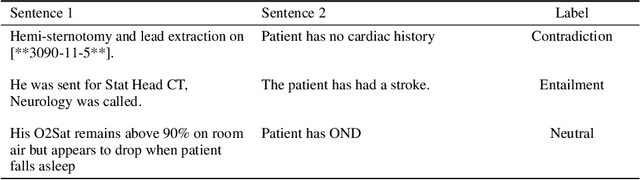
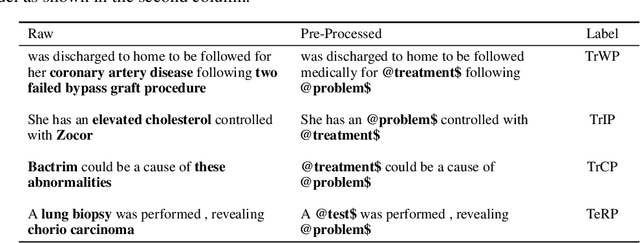
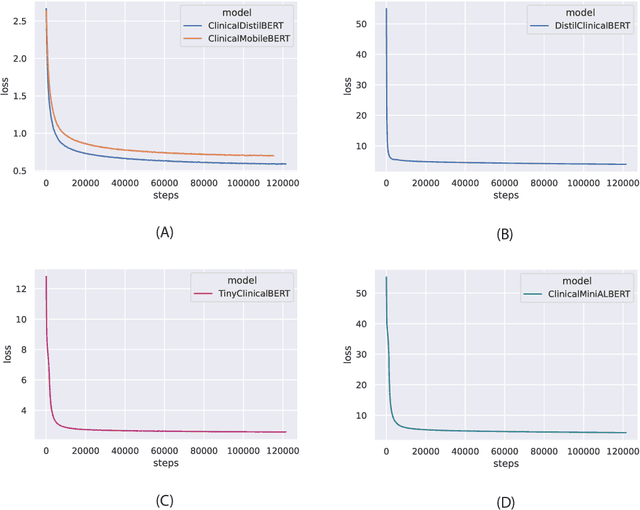
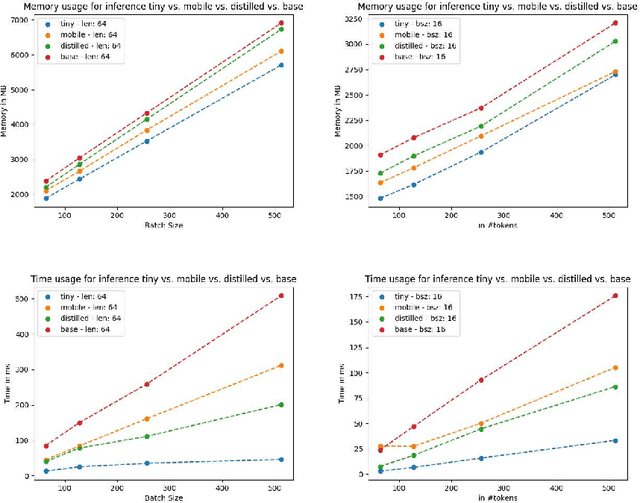
Abstract:Specialised pre-trained language models are becoming more frequent in NLP since they can potentially outperform models trained on generic texts. BioBERT and BioClinicalBERT are two examples of such models that have shown promise in medical NLP tasks. Many of these models are overparametrised and resource-intensive, but thanks to techniques like Knowledge Distillation (KD), it is possible to create smaller versions that perform almost as well as their larger counterparts. In this work, we specifically focus on development of compact language models for processing clinical texts (i.e. progress notes, discharge summaries etc). We developed a number of efficient lightweight clinical transformers using knowledge distillation and continual learning, with the number of parameters ranging from 15 million to 65 million. These models performed comparably to larger models such as BioBERT and ClinicalBioBERT and significantly outperformed other compact models trained on general or biomedical data. Our extensive evaluation was done across several standard datasets and covered a wide range of clinical text-mining tasks, including Natural Language Inference, Relation Extraction, Named Entity Recognition, and Sequence Classification. To our knowledge, this is the first comprehensive study specifically focused on creating efficient and compact transformers for clinical NLP tasks. The models and code used in this study can be found on our Huggingface profile at https://huggingface.co/nlpie and Github page at https://github.com/nlpie-research/Lightweight-Clinical-Transformers, respectively, promoting reproducibility of our results.
G-CMP: Graph-enhanced Contextual Matrix Profile for unsupervised anomaly detection in sensor-based remote health monitoring
Nov 29, 2022Abstract:Sensor-based remote health monitoring is used in industrial, urban and healthcare settings to monitor ongoing operation of equipment and human health. An important aim is to intervene early if anomalous events or adverse health is detected. In the wild, these anomaly detection approaches are challenged by noise, label scarcity, high dimensionality, explainability and wide variability in operating environments. The Contextual Matrix Profile (CMP) is a configurable 2-dimensional version of the Matrix Profile (MP) that uses the distance matrix of all subsequences of a time series to discover patterns and anomalies. The CMP is shown to enhance the effectiveness of the MP and other SOTA methods at detecting, visualising and interpreting true anomalies in noisy real world data from different domains. It excels at zooming out and identifying temporal patterns at configurable time scales. However, the CMP does not address cross-sensor information, and cannot scale to high dimensional data. We propose a novel, self-supervised graph-based approach for temporal anomaly detection that works on context graphs generated from the CMP distance matrix. The learned graph embeddings encode the anomalous nature of a time context. In addition, we evaluate other graph outlier algorithms for the same task. Given our pipeline is modular, graph construction, generation of graph embeddings, and pattern recognition logic can all be chosen based on the specific pattern detection application. We verified the effectiveness of graph-based anomaly detection and compared it with the CMP and 3 state-of-the art methods on two real-world healthcare datasets with different anomalies. Our proposed method demonstrated better recall, alert rate and generalisability.
MiniALBERT: Model Distillation via Parameter-Efficient Recursive Transformers
Oct 12, 2022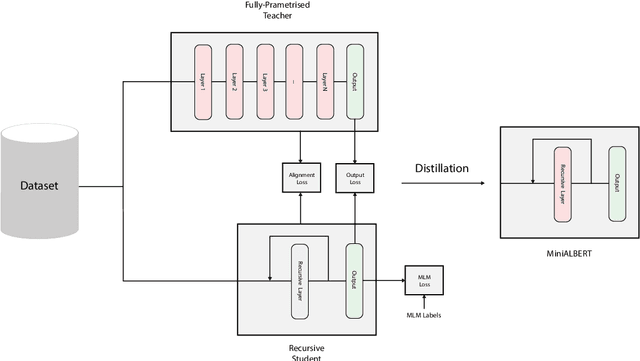
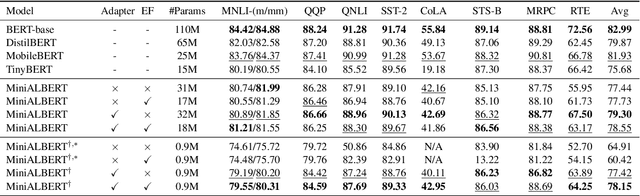
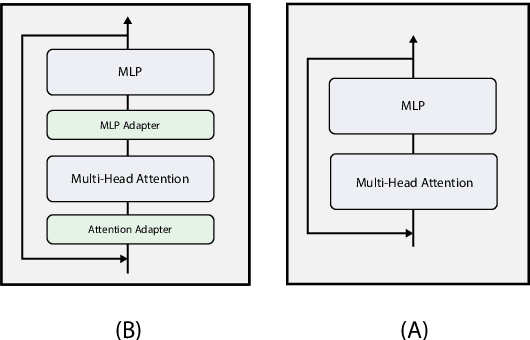
Abstract:Pre-trained Language Models (LMs) have become an integral part of Natural Language Processing (NLP) in recent years, due to their superior performance in downstream applications. In spite of this resounding success, the usability of LMs is constrained by computational and time complexity, along with their increasing size; an issue that has been referred to as `overparameterisation'. Different strategies have been proposed in the literature to alleviate these problems, with the aim to create effective compact models that nearly match the performance of their bloated counterparts with negligible performance losses. One of the most popular techniques in this area of research is model distillation. Another potent but underutilised technique is cross-layer parameter sharing. In this work, we combine these two strategies and present MiniALBERT, a technique for converting the knowledge of fully parameterised LMs (such as BERT) into a compact recursive student. In addition, we investigate the application of bottleneck adapters for layer-wise adaptation of our recursive student, and also explore the efficacy of adapter tuning for fine-tuning of compact models. We test our proposed models on a number of general and biomedical NLP tasks to demonstrate their viability and compare them with the state-of-the-art and other existing compact models. All the codes used in the experiments are available at https://github.com/nlpie-research/MiniALBERT. Our pre-trained compact models can be accessed from https://huggingface.co/nlpie.
On the Effectiveness of Compact Biomedical Transformers
Sep 07, 2022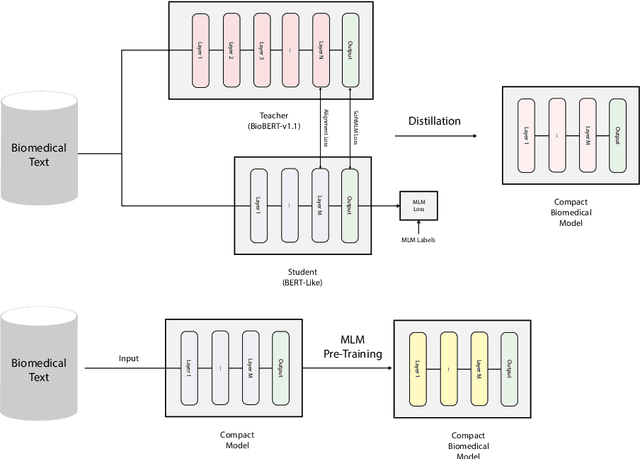
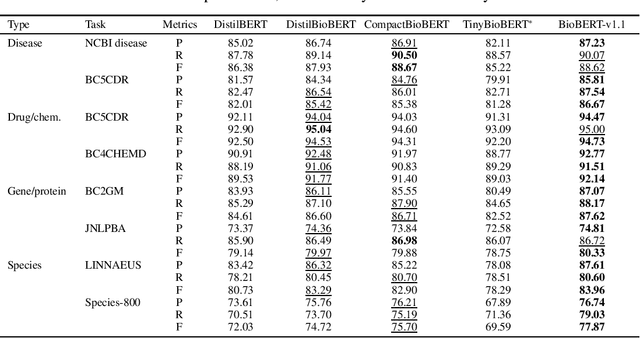
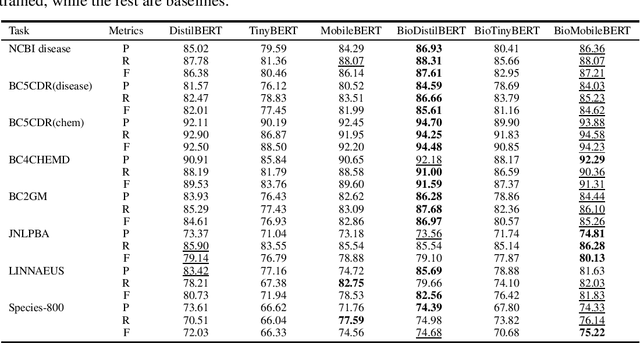
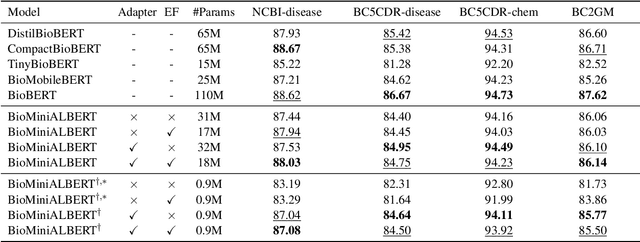
Abstract:Language models pre-trained on biomedical corpora, such as BioBERT, have recently shown promising results on downstream biomedical tasks. Many existing pre-trained models, on the other hand, are resource-intensive and computationally heavy owing to factors such as embedding size, hidden dimension, and number of layers. The natural language processing (NLP) community has developed numerous strategies to compress these models utilising techniques such as pruning, quantisation, and knowledge distillation, resulting in models that are considerably faster, smaller, and subsequently easier to use in practice. By the same token, in this paper we introduce six lightweight models, namely, BioDistilBERT, BioTinyBERT, BioMobileBERT, DistilBioBERT, TinyBioBERT, and CompactBioBERT which are obtained either by knowledge distillation from a biomedical teacher or continual learning on the Pubmed dataset via the Masked Language Modelling (MLM) objective. We evaluate all of our models on three biomedical tasks and compare them with BioBERT-v1.1 to create efficient lightweight models that perform on par with their larger counterparts. All the models will be publicly available on our Huggingface profile at https://huggingface.co/nlpie and the codes used to run the experiments will be available at https://github.com/nlpie-research/Compact-Biomedical-Transformers.
Privacy-aware Early Detection of COVID-19 through Adversarial Training
Jan 09, 2022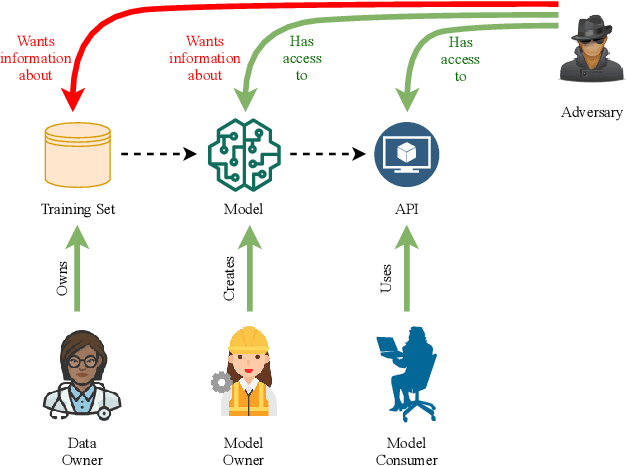
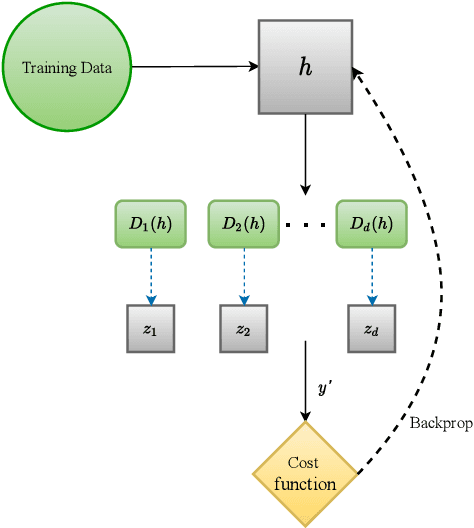

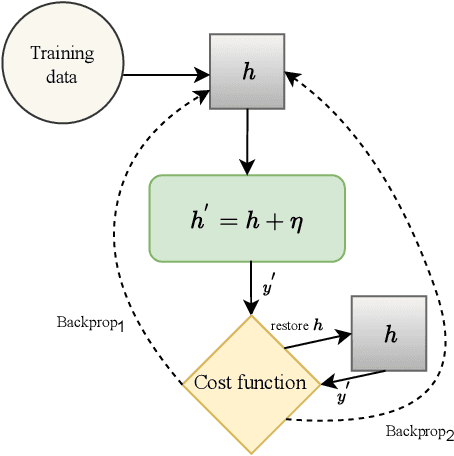
Abstract:Early detection of COVID-19 is an ongoing area of research that can help with triage, monitoring and general health assessment of potential patients and may reduce operational strain on hospitals that cope with the coronavirus pandemic. Different machine learning techniques have been used in the literature to detect coronavirus using routine clinical data (blood tests, and vital signs). Data breaches and information leakage when using these models can bring reputational damage and cause legal issues for hospitals. In spite of this, protecting healthcare models against leakage of potentially sensitive information is an understudied research area. In this work, we examine two machine learning approaches, intended to predict a patient's COVID-19 status using routinely collected and readily available clinical data. We employ adversarial training to explore robust deep learning architectures that protect attributes related to demographic information about the patients. The two models we examine in this work are intended to preserve sensitive information against adversarial attacks and information leakage. In a series of experiments using datasets from the Oxford University Hospitals, Bedfordshire Hospitals NHS Foundation Trust, University Hospitals Birmingham NHS Foundation Trust, and Portsmouth Hospitals University NHS Trust we train and test two neural networks that predict PCR test results using information from basic laboratory blood tests, and vital signs performed on a patients' arrival to hospital. We assess the level of privacy each one of the models can provide and show the efficacy and robustness of our proposed architectures against a comparable baseline. One of our main contributions is that we specifically target the development of effective COVID-19 detection models with built-in mechanisms in order to selectively protect sensitive attributes against adversarial attacks.
Designing A Clinically Applicable Deep Recurrent Model to Identify Neuropsychiatric Symptoms in People Living with Dementia Using In-Home Monitoring Data
Oct 19, 2021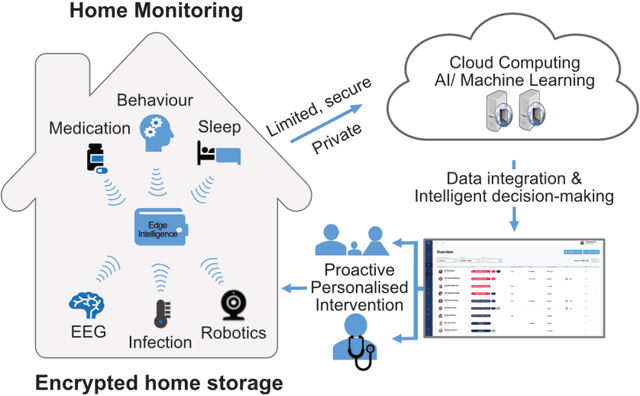
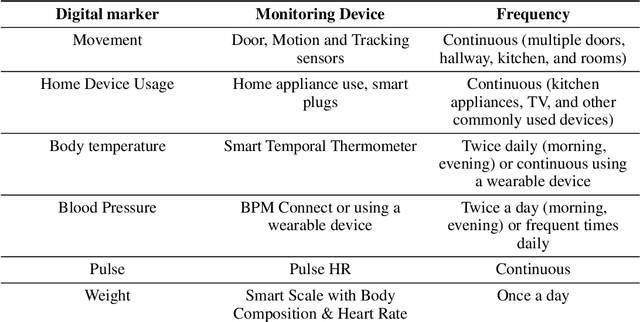

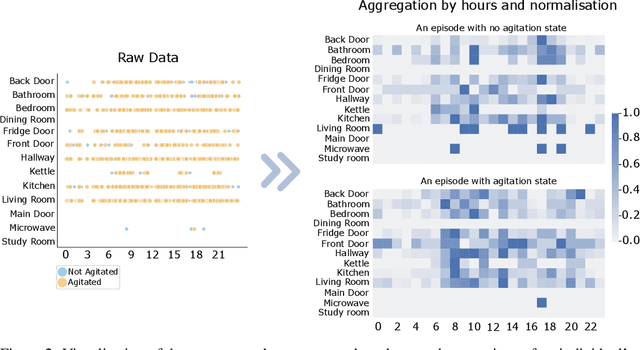
Abstract:Agitation is one of the neuropsychiatric symptoms with high prevalence in dementia which can negatively impact the Activities of Daily Living (ADL) and the independence of individuals. Detecting agitation episodes can assist in providing People Living with Dementia (PLWD) with early and timely interventions. Analysing agitation episodes will also help identify modifiable factors such as ambient temperature and sleep as possible components causing agitation in an individual. This preliminary study presents a supervised learning model to analyse the risk of agitation in PLWD using in-home monitoring data. The in-home monitoring data includes motion sensors, physiological measurements, and the use of kitchen appliances from 46 homes of PLWD between April 2019-June 2021. We apply a recurrent deep learning model to identify agitation episodes validated and recorded by a clinical monitoring team. We present the experiments to assess the efficacy of the proposed model. The proposed model achieves an average of 79.78% recall, 27.66% precision and 37.64% F1 scores when employing the optimal parameters, suggesting a good ability to recognise agitation events. We also discuss using machine learning models for analysing the behavioural patterns using continuous monitoring data and explore clinical applicability and the choices between sensitivity and specificity in-home monitoring applications.
Semi-supervised Learning for Identifying the Likelihood of Agitation in People with Dementia
May 14, 2021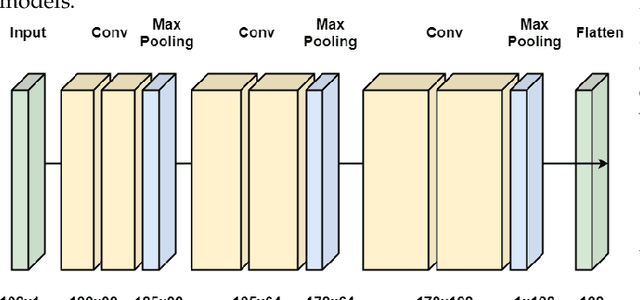
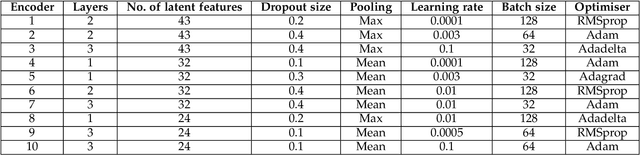
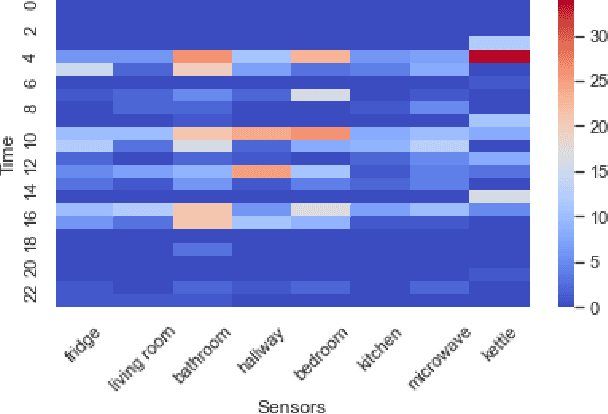
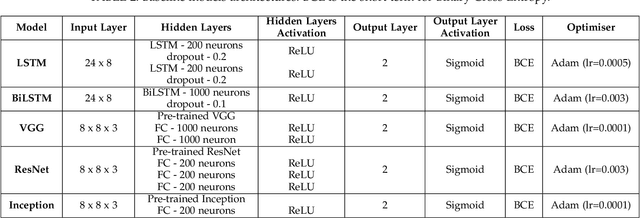
Abstract:Interpreting the environmental, behavioural and psychological data from in-home sensory observations and measurements can provide valuable insights into the health and well-being of individuals. Presents of neuropsychiatric and psychological symptoms in people with dementia have a significant impact on their well-being and disease prognosis. Agitation in people with dementia can be due to many reasons such as pain or discomfort, medical reasons such as side effects of a medicine, communication problems and environment. This paper discusses a model for analysing the risk of agitation in people with dementia and how in-home monitoring data can support them. We proposed a semi-supervised model which combines a self-supervised learning model and a Bayesian ensemble classification. We train and test the proposed model on a dataset from a clinical study. The dataset was collected from sensors deployed in 96 homes of patients with dementia. The proposed model outperforms the state-of-the-art models in recall and f1-score values by 20%. The model also indicates better generalisability compared to the baseline models.
A Hamiltonian Monte Carlo Model for Imputation and Augmentation of Healthcare Data
Mar 03, 2021



Abstract:Missing values exist in nearly all clinical studies because data for a variable or question are not collected or not available. Inadequate handling of missing values can lead to biased results and loss of statistical power in analysis. Existing models usually do not consider privacy concerns or do not utilise the inherent correlations across multiple features to impute the missing values. In healthcare applications, we are usually confronted with high dimensional and sometimes small sample size datasets that need more effective augmentation or imputation techniques. Besides, imputation and augmentation processes are traditionally conducted individually. However, imputing missing values and augmenting data can significantly improve generalisation and avoid bias in machine learning models. A Bayesian approach to impute missing values and creating augmented samples in high dimensional healthcare data is proposed in this work. We propose folded Hamiltonian Monte Carlo (F-HMC) with Bayesian inference as a more practical approach to process the cross-dimensional relations by applying a random walk and Hamiltonian dynamics to adapt posterior distribution and generate large-scale samples. The proposed method is applied to a cancer symptom assessment dataset and confirmed to enrich the quality of data in precision, accuracy, recall, F1 score, and propensity metric.
Bridging the Gap: Attending to Discontinuity in Identification of Multiword Expressions
Feb 27, 2019



Abstract:We introduce a new method to tag Multiword Expressions (MWEs) using a linguistically interpretable language-independent deep learning architecture. We specifically target discontinuity, an under-explored aspect that poses a significant challenge to computational treatment of MWEs. Two neural architectures are explored: Graph Convolutional Network (GCN) and multi-head self-attention. GCN leverages dependency parse information, and self-attention attends to long-range relations. We finally propose a combined model that integrates complementary information from both through a gating mechanism. The experiments on a standard multilingual dataset for verbal MWEs show that our model outperforms the baselines not only in the case of discontinuous MWEs but also in overall F-score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge