Sajad Darabi
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024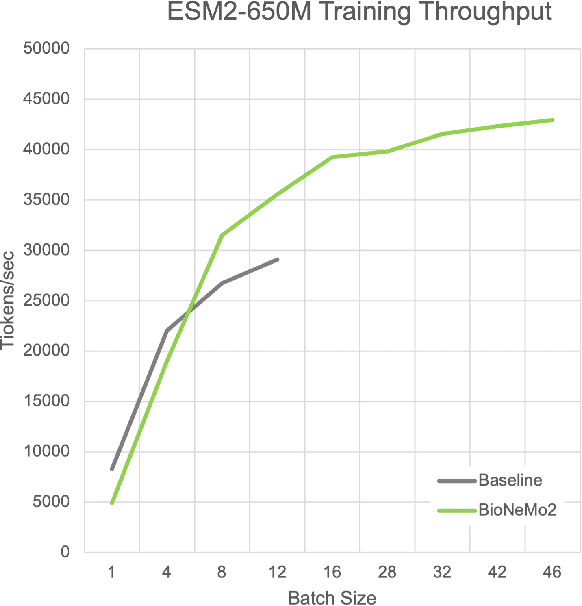
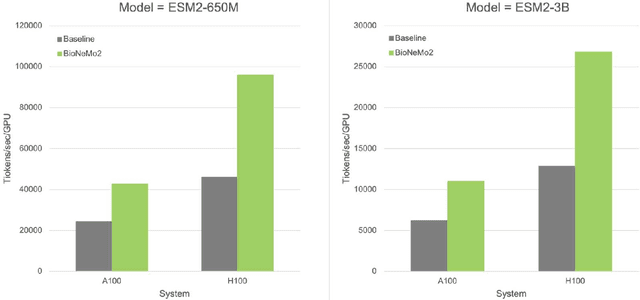
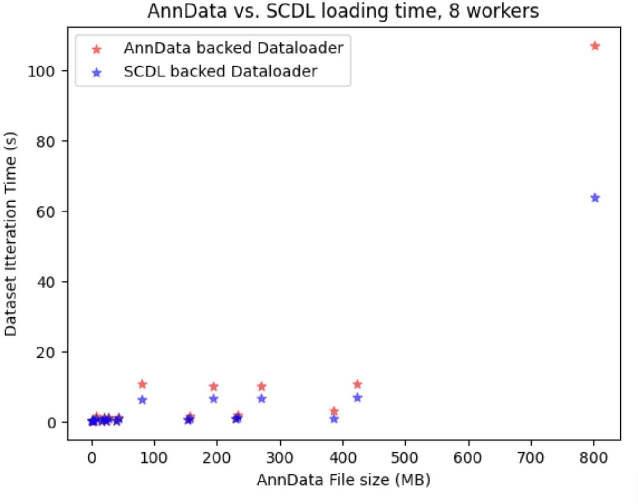
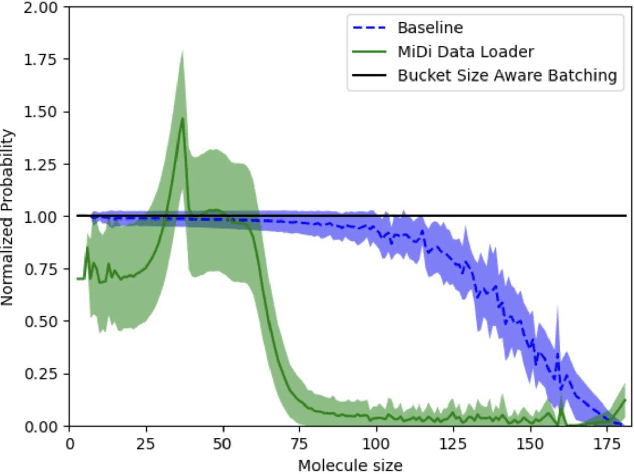
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
TSPP: A Unified Benchmarking Tool for Time-series Forecasting
Jan 08, 2024Abstract:While machine learning has witnessed significant advancements, the emphasis has largely been on data acquisition and model creation. However, achieving a comprehensive assessment of machine learning solutions in real-world settings necessitates standardization throughout the entire pipeline. This need is particularly acute in time series forecasting, where diverse settings impede meaningful comparisons between various methods. To bridge this gap, we propose a unified benchmarking framework that exposes the crucial modelling and machine learning decisions involved in developing time series forecasting models. This framework fosters seamless integration of models and datasets, aiding both practitioners and researchers in their development efforts. We benchmark recently proposed models within this framework, demonstrating that carefully implemented deep learning models with minimal effort can rival gradient-boosting decision trees requiring extensive feature engineering and expert knowledge.
Heterogenous Ensemble of Models for Molecular Property Prediction
Nov 20, 2022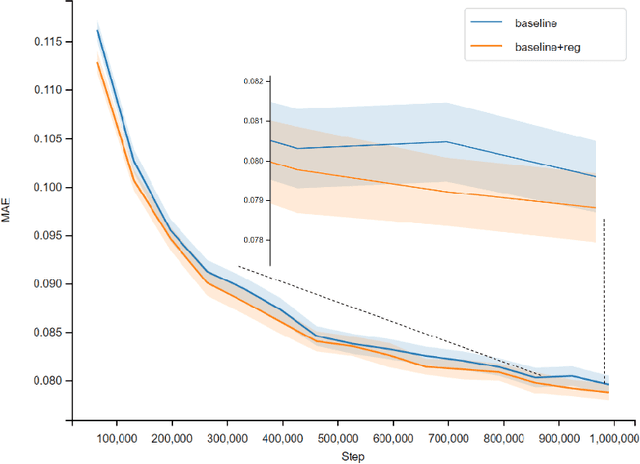
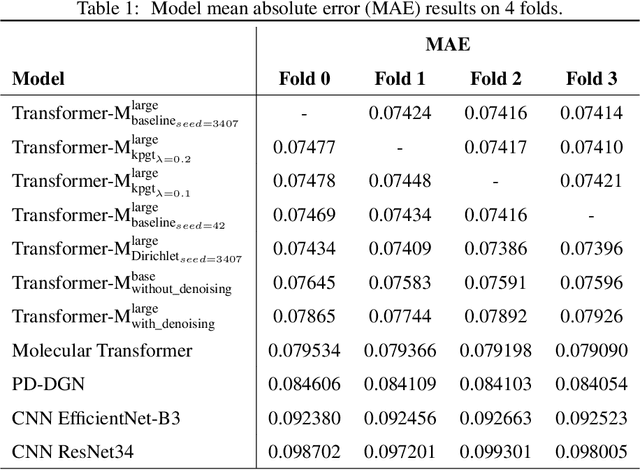
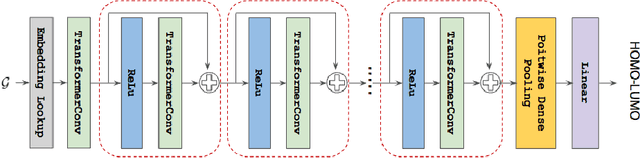
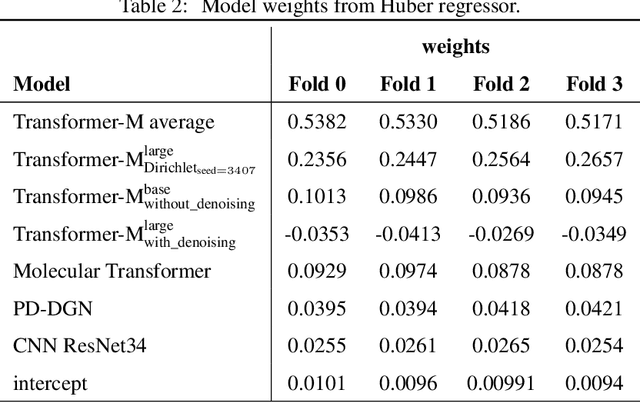
Abstract:Previous works have demonstrated the importance of considering different modalities on molecules, each of which provide a varied granularity of information for downstream property prediction tasks. Our method combines variants of the recent TransformerM architecture with Transformer, GNN, and ResNet backbone architectures. Models are trained on the 2D data, 3D data, and image modalities of molecular graphs. We ensemble these models with a HuberRegressor. The models are trained on 4 different train/validation splits of the original train + valid datasets. This yields a winning solution to the 2\textsuperscript{nd} edition of the OGB Large-Scale Challenge (2022) on the PCQM4Mv2 molecular property prediction dataset. Our proposed method achieves a test-challenge MAE of $0.0723$ and a validation MAE of $0.07145$. Total inference time for our solution is less than 2 hours. We open-source our code at https://github.com/jfpuget/NVIDIA-PCQM4Mv2.
A Framework for Large Scale Synthetic Graph Dataset Generation
Oct 06, 2022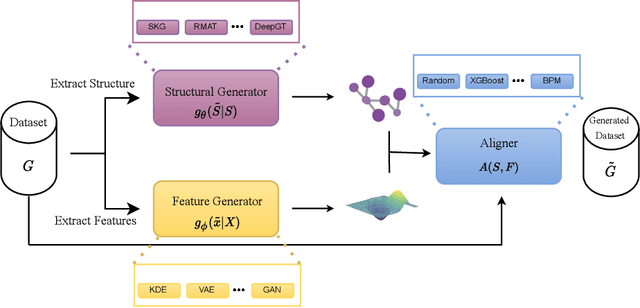
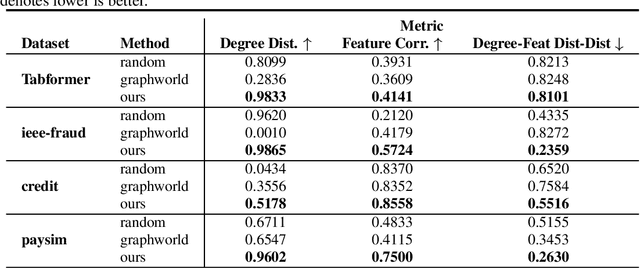
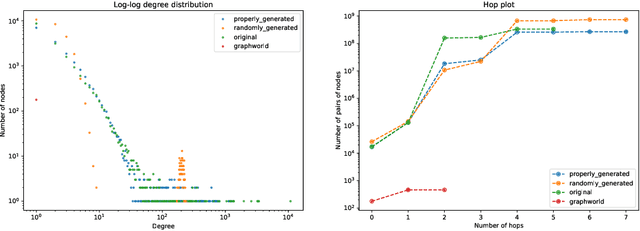
Abstract:Recently there has been increasing interest in developing and deploying deep graph learning algorithms for many graph analysis tasks such as node and edge classification, link prediction, and clustering with numerous practical applications such as fraud detection, drug discovery, or recommender systems. Allbeit there is a limited number of publicly available graph-structured datasets, most of which are tiny compared to production-sized applications with trillions of edges and billions of nodes. Further, new algorithms and models are benchmarked across similar datasets with similar properties. In this work, we tackle this shortcoming by proposing a scalable synthetic graph generation tool that can mimic the original data distribution of real-world graphs and scale them to arbitrary sizes. This tool can be used then to learn a set of parametric models from proprietary datasets that can subsequently be released to researchers to study various graph methods on the synthetic data increasing prototype development and novel applications. Finally, the performance of the graph learning algorithms depends not only on the size but also on the dataset's structure. We show how our framework generalizes across a set of datasets, mimicking both structural and feature distributions as well as its scalability across varying dataset sizes.
Contrastive Mixup: Self- and Semi-Supervised learning for Tabular Domain
Sep 01, 2021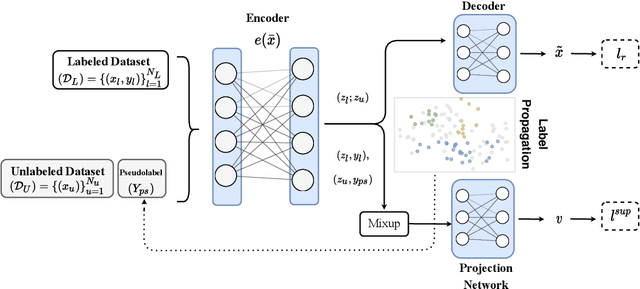
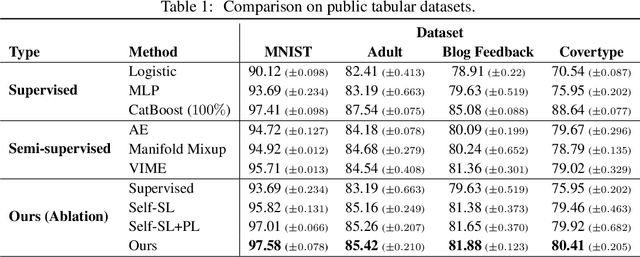
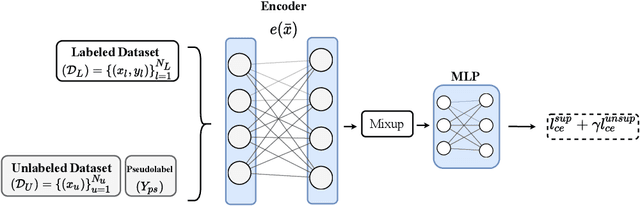
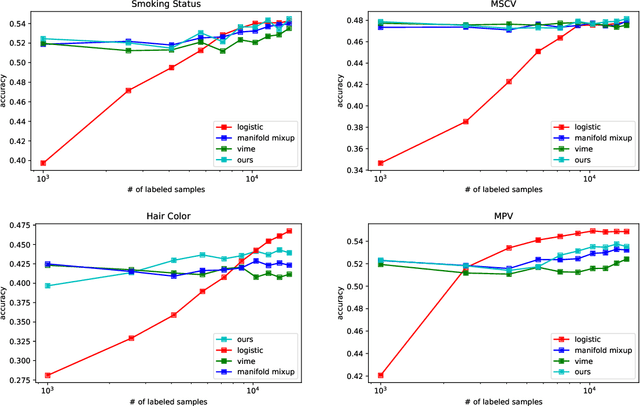
Abstract:Recent literature in self-supervised has demonstrated significant progress in closing the gap between supervised and unsupervised methods in the image and text domains. These methods rely on domain-specific augmentations that are not directly amenable to the tabular domain. Instead, we introduce Contrastive Mixup, a semi-supervised learning framework for tabular data and demonstrate its effectiveness in limited annotated data settings. Our proposed method leverages Mixup-based augmentation under the manifold assumption by mapping samples to a low dimensional latent space and encourage interpolated samples to have high a similarity within the same labeled class. Unlabeled samples are additionally employed via a transductive label propagation method to further enrich the set of similar and dissimilar pairs that can be used in the contrastive loss term. We demonstrate the effectiveness of the proposed framework on public tabular datasets and real-world clinical datasets.
Synthesising Multi-Modal Minority Samples for Tabular Data
May 17, 2021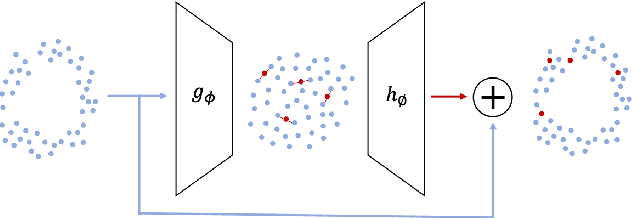
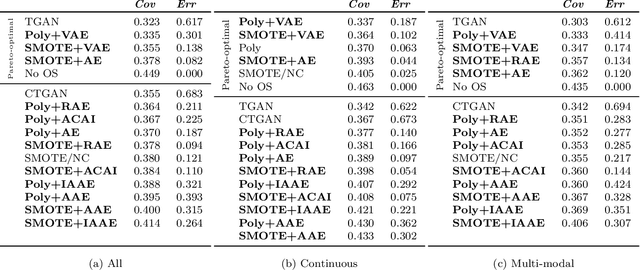
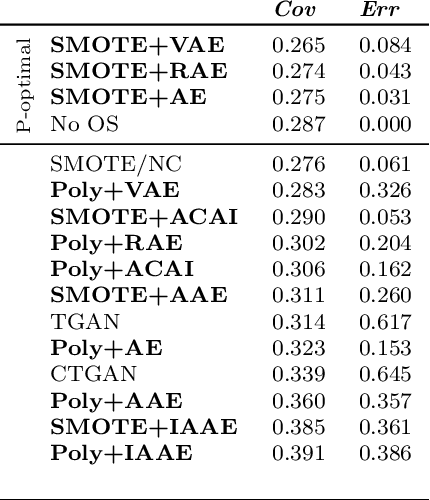
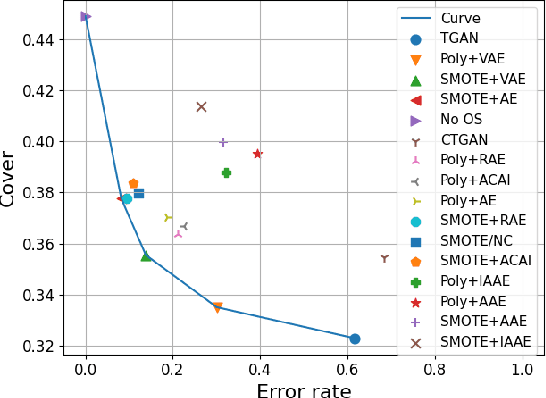
Abstract:Real-world binary classification tasks are in many cases imbalanced, where the minority class is much smaller than the majority class. This skewness is challenging for machine learning algorithms as they tend to focus on the majority and greatly misclassify the minority. Adding synthetic minority samples to the dataset before training the model is a popular technique to address this difficulty and is commonly achieved by interpolating minority samples. Tabular datasets are often multi-modal and contain discrete (categorical) features in addition to continuous ones which makes interpolation of samples non-trivial. To address this, we propose a latent space interpolation framework which (1) maps the multi-modal samples to a dense continuous latent space using an autoencoder; (2) applies oversampling by interpolation in the latent space; and (3) maps the synthetic samples back to the original feature space. We defined metrics to directly evaluate the quality of the minority data generated and showed that our framework generates better synthetic data than the existing methods. Furthermore, the superior synthetic data yields better prediction quality in downstream binary classification tasks, as was demonstrated in extensive experiments with 27 publicly available real-world datasets
Group-Connected Multilayer Perceptron Networks
Dec 20, 2019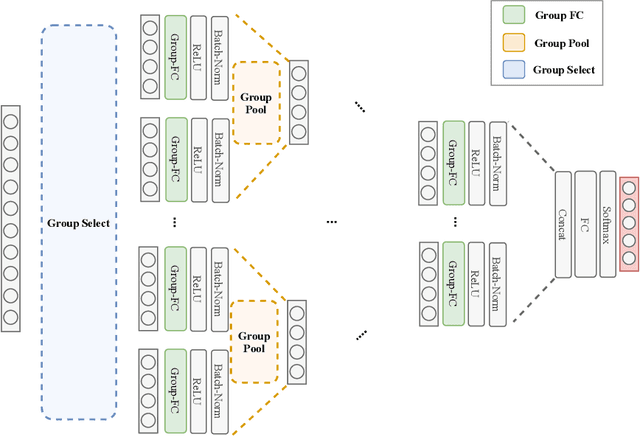
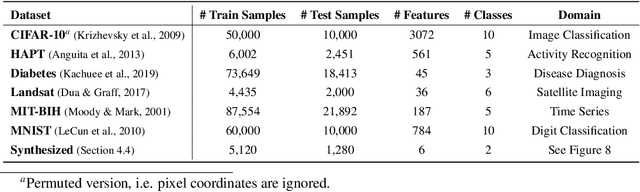
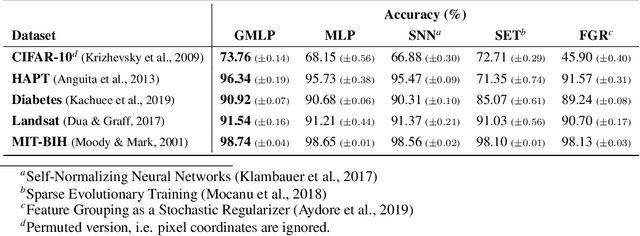
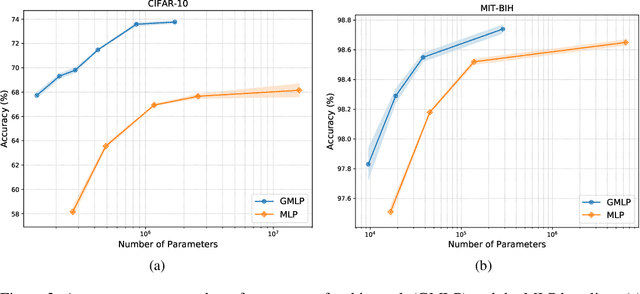
Abstract:Despite the success of deep learning in domains such as image, voice, and graphs, there has been little progress in deep representation learning for domains without a known structure between features. For instance, a tabular dataset of different demographic and clinical factors where the feature interactions are not given as a prior. In this paper, we propose Group-Connected Multilayer Perceptron (GMLP) networks to enable deep representation learning in these domains. GMLP is based on the idea of learning expressive feature combinations (groups) and exploiting them to reduce the network complexity by defining local group-wise operations. During the training phase, GMLP learns a sparse feature grouping matrix using temperature annealing softmax with an added entropy loss term to encourage the sparsity. Furthermore, an architecture is suggested which resembles binary trees, where group-wise operations are followed by pooling operations to combine information; reducing the number of groups as the network grows in depth. To evaluate the proposed method, we conducted experiments on five different real-world datasets covering various application areas. Additionally, we provide visualizations on MNIST and synthesized data. According to the results, GMLP is able to successfully learn and exploit expressive feature combinations and achieve state-of-the-art classification performance on different datasets.
Unsupervised Representation for EHR Signals and Codes as Patient Status Vector
Oct 04, 2019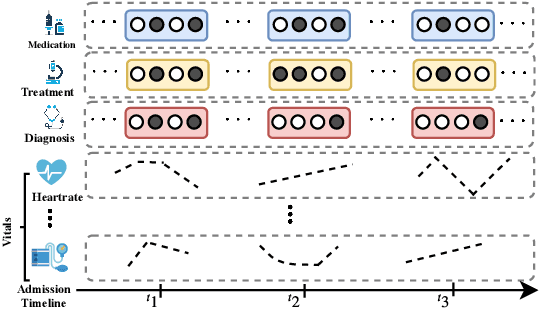
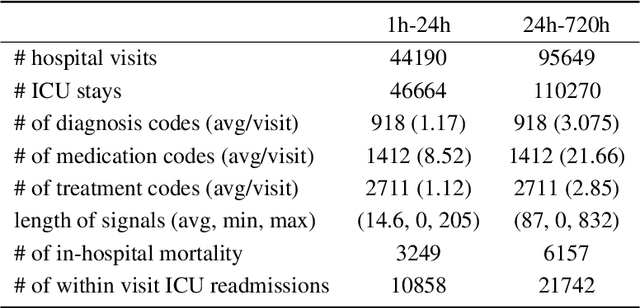
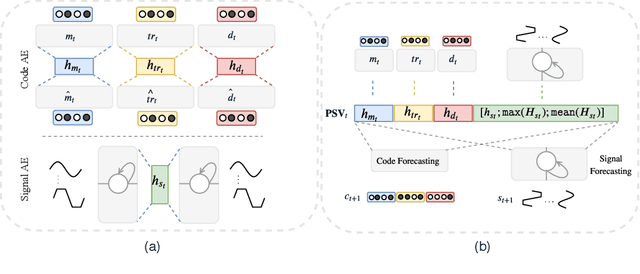
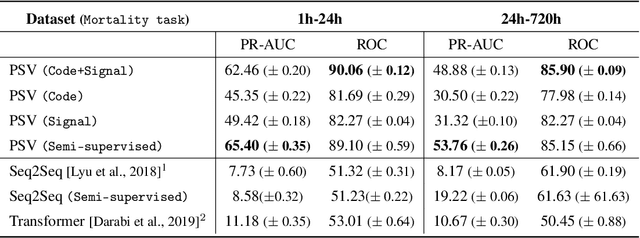
Abstract:Effective modeling of electronic health records presents many challenges as they contain large amounts of irregularity most of which are due to the varying procedures and diagnosis a patient may have. Despite the recent progress in machine learning, unsupervised learning remains largely at open, especially in the healthcare domain. In this work, we present a two-step unsupervised representation learning scheme to summarize the multi-modal clinical time series consisting of signals and medical codes into a patient status vector. First, an auto-encoder step is used to reduce sparse medical codes and clinical time series into a distributed representation. Subsequently, the concatenation of the distributed representations is further fine-tuned using a forecasting task. We evaluate the usefulness of the representation on two downstream tasks: mortality and readmission. Our proposed method shows improved generalization performance for both short duration ICU visits and long duration ICU visits.
TAPER: Time-Aware Patient EHR Representation
Aug 16, 2019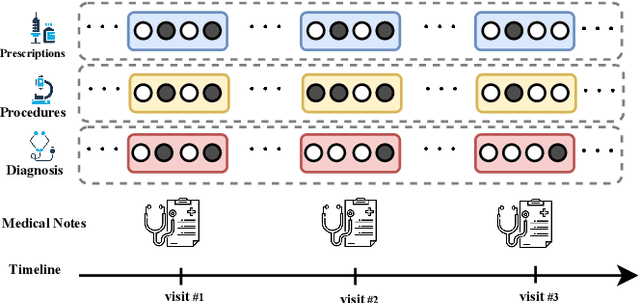
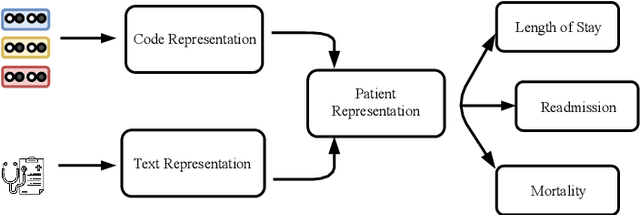
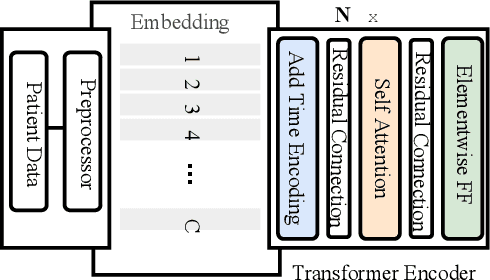
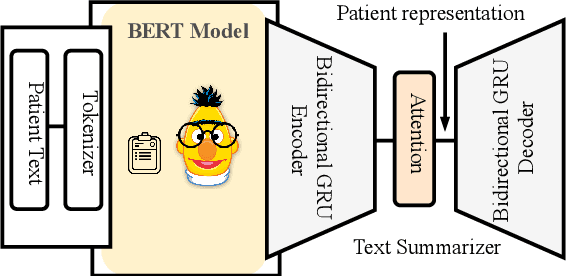
Abstract:Effective representation learning of electronic health records is a challenging task and is becoming more important as the availability of such data is becoming pervasive. The data contained in these records are irregular and contain multiple modalities such as notes, and medical codes. They are preempted by medical conditions the patient may have, and are typically jotted down by medical staff. Accompanying codes are notes containing valuable information about patients beyond the structured information contained in electronic health records. We use transformer networks and the recently proposed BERT language model to embed these data streams into a unified vector representation. The presented approach effectively encodes a patient's visit data into a single distributed representation, which can be used for downstream tasks. Our model demonstrates superior performance and generalization on mortality, readmission and length of stay tasks using the publicly available MIMIC-III ICU dataset.
Generative Imputation and Stochastic Prediction
May 22, 2019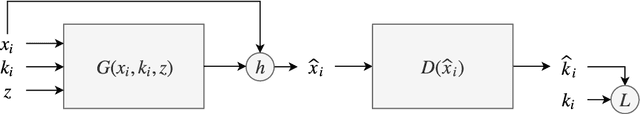
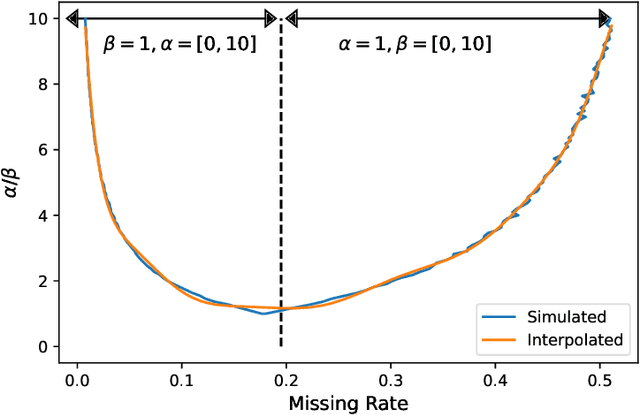
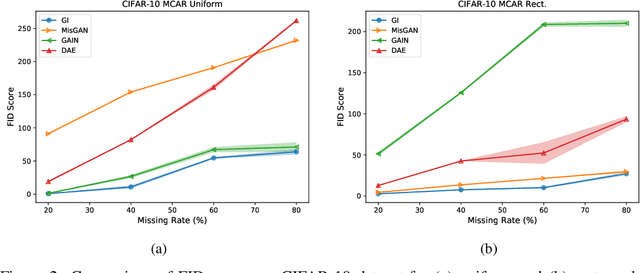
Abstract:In many machine learning applications, we are faced with incomplete datasets. In the literature, missing data imputation techniques have been mostly concerned with filling missing values. However, the existence of missing values is synonymous with uncertainties not only over the distribution of missing values but also over target class assignments that require careful consideration. The objectives of this paper are twofold. First, we proposed a method for generating imputations from the conditional distribution of missing values given observed values. Second, we use the generated samples to estimate the distribution of target assignments given incomplete data. In order to generate imputations, we train a simple and effective generator network to generate imputations that a discriminator network is tasked to distinguish. Following this, a predictor network is trained using imputed samples from the generator network to capture the classification uncertainties and make predictions accordingly. The proposed method is evaluated on CIFAR-10 image dataset as well as two real-world tabular classification datasets, under various missingness rates and structures. Our experimental results show the effectiveness of the proposed method in generating imputations, as well as providing estimates for the class uncertainties in a classification task when faced with missing values.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge