Shayan Fazeli
A Self-supervised Framework for Improved Data-Driven Monitoring of Stress via Multi-modal Passive Sensing
Mar 24, 2023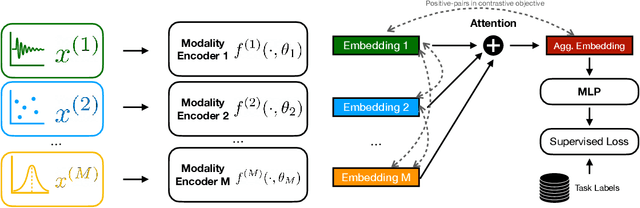

Abstract:Recent advances in remote health monitoring systems have significantly benefited patients and played a crucial role in improving their quality of life. However, while physiological health-focused solutions have demonstrated increasing success and maturity, mental health-focused applications have seen comparatively limited success in spite of the fact that stress and anxiety disorders are among the most common issues people deal with in their daily lives. In the hopes of furthering progress in this domain through the development of a more robust analytic framework for the measurement of indicators of mental health, we propose a multi-modal semi-supervised framework for tracking physiological precursors of the stress response. Our methodology enables utilizing multi-modal data of differing domains and resolutions from wearable devices and leveraging them to map short-term episodes to semantically efficient embeddings for a given task. Additionally, we leverage an inter-modality contrastive objective, with the advantages of rendering our framework both modular and scalable. The focus on optimizing both local and global aspects of our embeddings via a hierarchical structure renders transferring knowledge and compatibility with other devices easier to achieve. In our pipeline, a task-specific pooling based on an attention mechanism, which estimates the contribution of each modality on an instance level, computes the final embeddings for observations. This additionally provides a thorough diagnostic insight into the data characteristics and highlights the importance of signals in the broader view of predicting episodes annotated per mental health status. We perform training experiments using a corpus of real-world data on perceived stress, and our results demonstrate the efficacy of the proposed approach in performance improvements.
Heterogenous Ensemble of Models for Molecular Property Prediction
Nov 20, 2022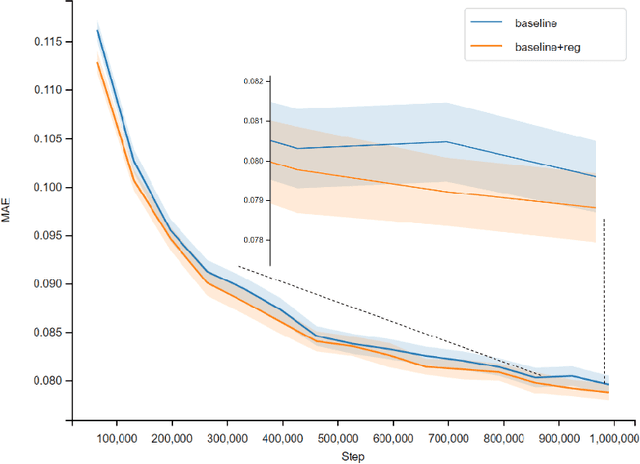
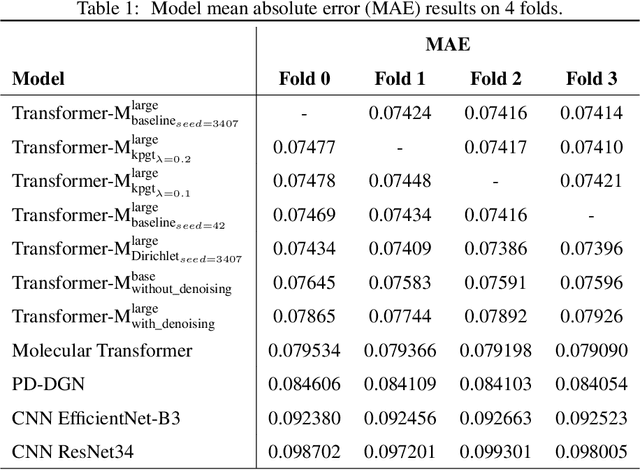
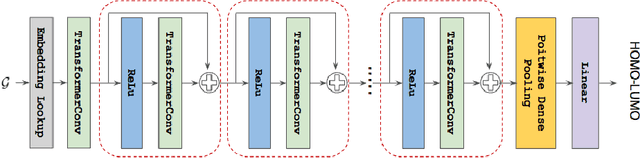
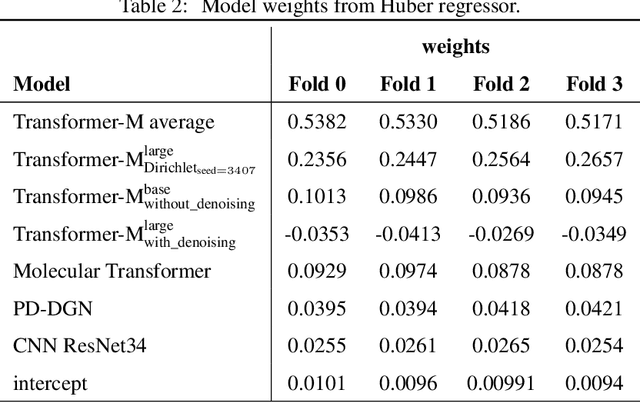
Abstract:Previous works have demonstrated the importance of considering different modalities on molecules, each of which provide a varied granularity of information for downstream property prediction tasks. Our method combines variants of the recent TransformerM architecture with Transformer, GNN, and ResNet backbone architectures. Models are trained on the 2D data, 3D data, and image modalities of molecular graphs. We ensemble these models with a HuberRegressor. The models are trained on 4 different train/validation splits of the original train + valid datasets. This yields a winning solution to the 2\textsuperscript{nd} edition of the OGB Large-Scale Challenge (2022) on the PCQM4Mv2 molecular property prediction dataset. Our proposed method achieves a test-challenge MAE of $0.0723$ and a validation MAE of $0.07145$. Total inference time for our solution is less than 2 hours. We open-source our code at https://github.com/jfpuget/NVIDIA-PCQM4Mv2.
Beyond Labels: Visual Representations for Bone Marrow Cell Morphology Recognition
May 19, 2022



Abstract:Analyzing and inspecting bone marrow cell cytomorphology is a critical but highly complex and time-consuming component of hematopathology diagnosis. Recent advancements in artificial intelligence have paved the way for the application of deep learning algorithms to complex medical tasks. Nevertheless, there are many challenges in applying effective learning algorithms to medical image analysis, such as the lack of sufficient and reliably annotated training datasets and the highly class-imbalanced nature of most medical data. Here, we improve on the state-of-the-art methodologies of bone marrow cell recognition by deviating from sole reliance on labeled data and leveraging self-supervision in training our learning models. We investigate our approach's effectiveness in identifying bone marrow cell types. Our experiments demonstrate significant performance improvements in conducting different bone marrow cell recognition tasks compared to the current state-of-the-art methodologies.
Contrastive Mixup: Self- and Semi-Supervised learning for Tabular Domain
Sep 01, 2021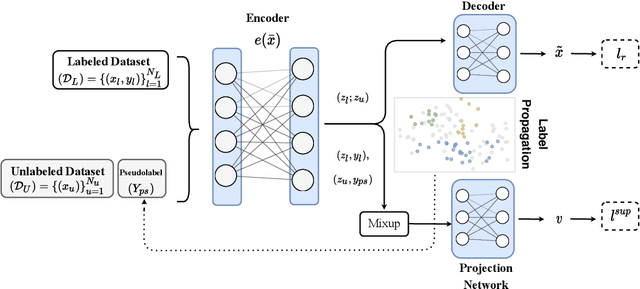
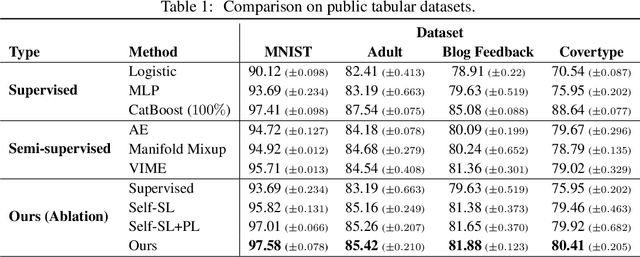
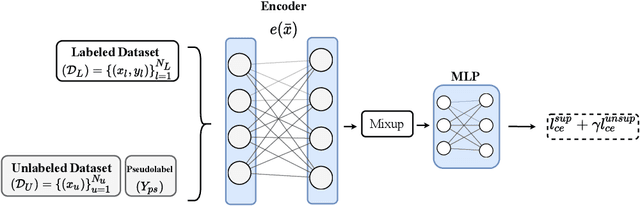
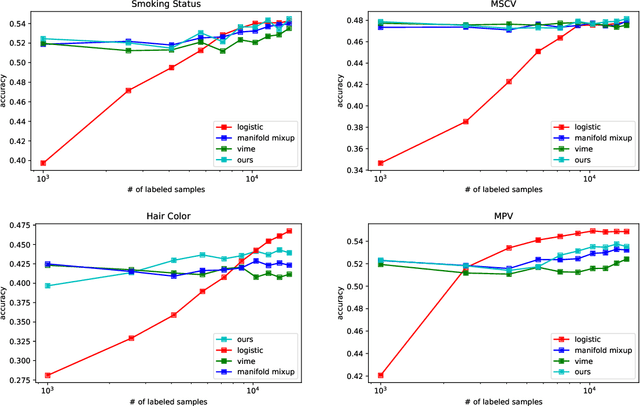
Abstract:Recent literature in self-supervised has demonstrated significant progress in closing the gap between supervised and unsupervised methods in the image and text domains. These methods rely on domain-specific augmentations that are not directly amenable to the tabular domain. Instead, we introduce Contrastive Mixup, a semi-supervised learning framework for tabular data and demonstrate its effectiveness in limited annotated data settings. Our proposed method leverages Mixup-based augmentation under the manifold assumption by mapping samples to a low dimensional latent space and encourage interpolated samples to have high a similarity within the same labeled class. Unlabeled samples are additionally employed via a transductive label propagation method to further enrich the set of similar and dissimilar pairs that can be used in the contrastive loss term. We demonstrate the effectiveness of the proposed framework on public tabular datasets and real-world clinical datasets.
A Framework for Neural Topic Modeling of Text Corpora
Aug 19, 2021

Abstract:Topic Modeling refers to the problem of discovering the main topics that have occurred in corpora of textual data, with solutions finding crucial applications in numerous fields. In this work, inspired by the recent advancements in the Natural Language Processing domain, we introduce FAME, an open-source framework enabling an efficient mechanism of extracting and incorporating textual features and utilizing them in discovering topics and clustering text documents that are semantically similar in a corpus. These features range from traditional approaches (e.g., frequency-based) to the most recent auto-encoding embeddings from transformer-based language models such as BERT model family. To demonstrate the effectiveness of this library, we conducted experiments on the well-known News-Group dataset. The library is available online.
Unsupervised Acute Intracranial Hemorrhage Segmentation with Mixture Models
May 12, 2021



Abstract:Intracranial hemorrhage occurs when blood vessels rupture or leak within the brain tissue or elsewhere inside the skull. It can be caused by physical trauma or by various medical conditions and in many cases leads to death. The treatment must be started as soon as possible, and therefore the hemorrhage should be diagnosed accurately and quickly. The diagnosis is usually performed by a radiologist who analyses a Computed Tomography (CT) scan containing a large number of cross-sectional images throughout the brain. Analysing each image manually can be very time-consuming, but automated techniques can help speed up the process. While much of the recent research has focused on solving this problem by using supervised machine learning algorithms, publicly-available training data remains scarce due to privacy concerns. This problem can be alleviated by unsupervised algorithms. In this paper, we propose a fully-unsupervised algorithm which is based on the mixture models. Our algorithm utilizes the fact that the properties of hemorrhage and healthy tissues follow different distributions, and therefore an appropriate formulation of these distributions allows us to separate them through an Expectation-Maximization process. In addition, our algorithm is able to adaptively determine the number of clusters such that all the hemorrhage regions can be found without including noisy voxels. We demonstrate the results of our algorithm on publicly-available datasets that contain all different hemorrhage types in various sizes and intensities, and our results are compared to earlier unsupervised and supervised algorithms. The results show that our algorithm can outperform the other algorithms with most hemorrhage types.
COVID-19 and Big Data: Multi-faceted Analysis for Spatio-temporal Understanding of the Pandemic with Social Media Conversations
Apr 22, 2021Abstract:COVID-19 has been devastating the world since the end of 2019 and has continued to play a significant role in major national and worldwide events, and consequently, the news. In its wake, it has left no life unaffected. Having earned the world's attention, social media platforms have served as a vehicle for the global conversation about COVID-19. In particular, many people have used these sites in order to express their feelings, experiences, and observations about the pandemic. We provide a multi-faceted analysis of critical properties exhibited by these conversations on social media regarding the novel coronavirus pandemic. We present a framework for analysis, mining, and tracking the critical content and characteristics of social media conversations around the pandemic. Focusing on Twitter and Reddit, we have gathered a large-scale dataset on COVID-19 social media conversations. Our analyses cover tracking potential reports on virus acquisition, symptoms, conversation topics, and language complexity measures through time and by region across the United States. We also present a BERT-based model for recognizing instances of hateful tweets in COVID-19 conversations, which achieves a lower error-rate than the state-of-the-art performance. Our results provide empirical validation for the effectiveness of our proposed framework and further demonstrate that social media data can be efficiently leveraged to provide public health experts with inexpensive but thorough insight over the course of an outbreak.
A Flexible and Intelligent Framework for Remote Health Monitoring Dashboards
Jun 09, 2020


Abstract:Developing and maintaining monitoring panels is undoubtedly the main task in the remote patient monitoring (RPM) systems. Due to the significant variations in desired functionalities, data sources, and objectives, designing an efficient dashboard that responds to the various needs in an RPM project is generally a cumbersome task to carry out. In this work, we present ViSierra, a framework for designing data monitoring dashboards in RPM projects. The abstractions and different components of this open-source project are explained, and examples are provided to support our claim concerning the effectiveness of this framework in preparing fast, efficient, and accurate monitoring platforms with minimal coding. These platforms will cover all the necessary aspects in a traditional RPM project and combine them with novel functionalities such as machine learning solutions and provide better data analysis instruments for the experts to track the information.
Group-Connected Multilayer Perceptron Networks
Dec 20, 2019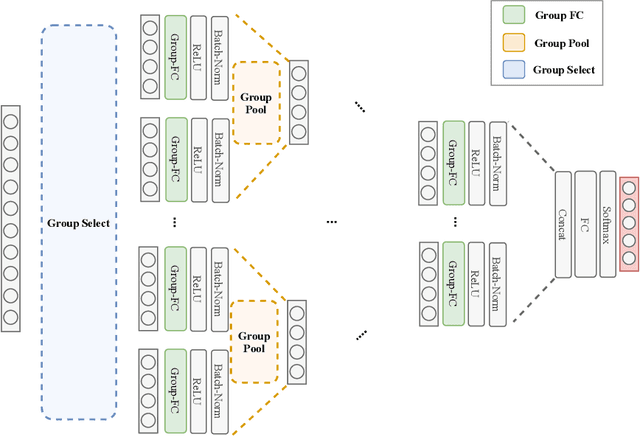
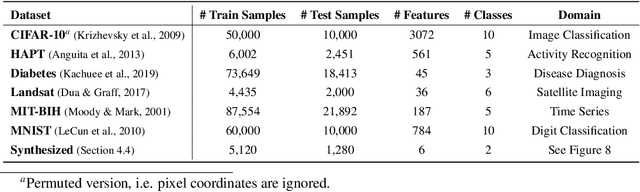
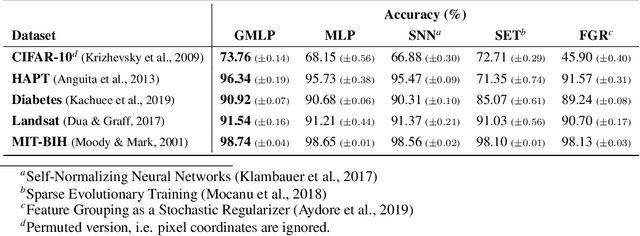
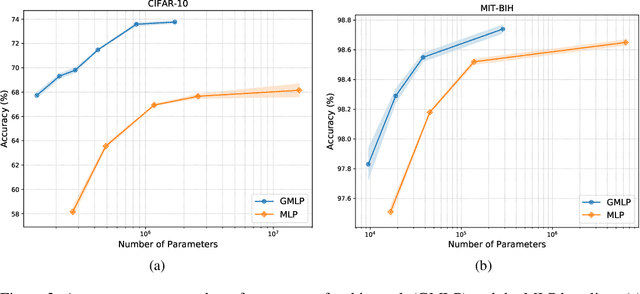
Abstract:Despite the success of deep learning in domains such as image, voice, and graphs, there has been little progress in deep representation learning for domains without a known structure between features. For instance, a tabular dataset of different demographic and clinical factors where the feature interactions are not given as a prior. In this paper, we propose Group-Connected Multilayer Perceptron (GMLP) networks to enable deep representation learning in these domains. GMLP is based on the idea of learning expressive feature combinations (groups) and exploiting them to reduce the network complexity by defining local group-wise operations. During the training phase, GMLP learns a sparse feature grouping matrix using temperature annealing softmax with an added entropy loss term to encourage the sparsity. Furthermore, an architecture is suggested which resembles binary trees, where group-wise operations are followed by pooling operations to combine information; reducing the number of groups as the network grows in depth. To evaluate the proposed method, we conducted experiments on five different real-world datasets covering various application areas. Additionally, we provide visualizations on MNIST and synthesized data. According to the results, GMLP is able to successfully learn and exploit expressive feature combinations and achieve state-of-the-art classification performance on different datasets.
TAPER: Time-Aware Patient EHR Representation
Aug 16, 2019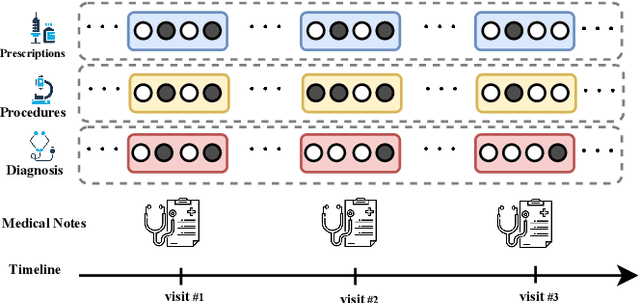
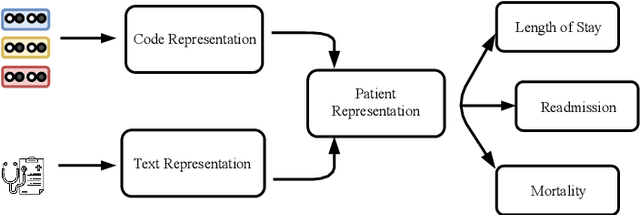
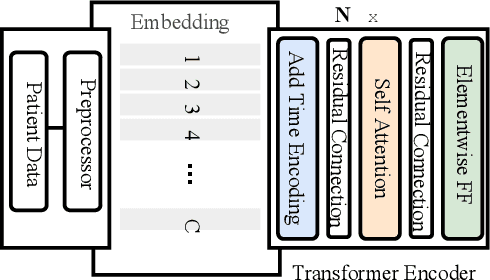
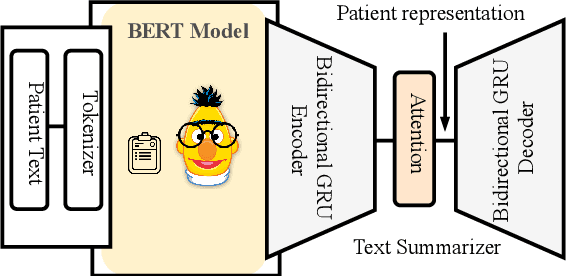
Abstract:Effective representation learning of electronic health records is a challenging task and is becoming more important as the availability of such data is becoming pervasive. The data contained in these records are irregular and contain multiple modalities such as notes, and medical codes. They are preempted by medical conditions the patient may have, and are typically jotted down by medical staff. Accompanying codes are notes containing valuable information about patients beyond the structured information contained in electronic health records. We use transformer networks and the recently proposed BERT language model to embed these data streams into a unified vector representation. The presented approach effectively encodes a patient's visit data into a single distributed representation, which can be used for downstream tasks. Our model demonstrates superior performance and generalization on mortality, readmission and length of stay tasks using the publicly available MIMIC-III ICU dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge