Alexandre Milesi
NVIDIA Nemotron 3: Efficient and Open Intelligence
Dec 24, 2025Abstract:We introduce the Nemotron 3 family of models - Nano, Super, and Ultra. These models deliver strong agentic, reasoning, and conversational capabilities. The Nemotron 3 family uses a Mixture-of-Experts hybrid Mamba-Transformer architecture to provide best-in-class throughput and context lengths of up to 1M tokens. Super and Ultra models are trained with NVFP4 and incorporate LatentMoE, a novel approach that improves model quality. The two larger models also include MTP layers for faster text generation. All Nemotron 3 models are post-trained using multi-environment reinforcement learning enabling reasoning, multi-step tool use, and support granular reasoning budget control. Nano, the smallest model, outperforms comparable models in accuracy while remaining extremely cost-efficient for inference. Super is optimized for collaborative agents and high-volume workloads such as IT ticket automation. Ultra, the largest model, provides state-of-the-art accuracy and reasoning performance. Nano is released together with its technical report and this white paper, while Super and Ultra will follow in the coming months. We will openly release the model weights, pre- and post-training software, recipes, and all data for which we hold redistribution rights.
Nemotron 3 Nano: Open, Efficient Mixture-of-Experts Hybrid Mamba-Transformer Model for Agentic Reasoning
Dec 23, 2025Abstract:We present Nemotron 3 Nano 30B-A3B, a Mixture-of-Experts hybrid Mamba-Transformer language model. Nemotron 3 Nano was pretrained on 25 trillion text tokens, including more than 3 trillion new unique tokens over Nemotron 2, followed by supervised fine tuning and large-scale RL on diverse environments. Nemotron 3 Nano achieves better accuracy than our previous generation Nemotron 2 Nano while activating less than half of the parameters per forward pass. It achieves up to 3.3x higher inference throughput than similarly-sized open models like GPT-OSS-20B and Qwen3-30B-A3B-Thinking-2507, while also being more accurate on popular benchmarks. Nemotron 3 Nano demonstrates enhanced agentic, reasoning, and chat abilities and supports context lengths up to 1M tokens. We release both our pretrained Nemotron 3 Nano 30B-A3B Base and post-trained Nemotron 3 Nano 30B-A3B checkpoints on Hugging Face.
NVIDIA Nemotron Nano V2 VL
Nov 07, 2025Abstract:We introduce Nemotron Nano V2 VL, the latest model of the Nemotron vision-language series designed for strong real-world document understanding, long video comprehension, and reasoning tasks. Nemotron Nano V2 VL delivers significant improvements over our previous model, Llama-3.1-Nemotron-Nano-VL-8B, across all vision and text domains through major enhancements in model architecture, datasets, and training recipes. Nemotron Nano V2 VL builds on Nemotron Nano V2, a hybrid Mamba-Transformer LLM, and innovative token reduction techniques to achieve higher inference throughput in long document and video scenarios. We are releasing model checkpoints in BF16, FP8, and FP4 formats and sharing large parts of our datasets, recipes and training code.
Heterogenous Ensemble of Models for Molecular Property Prediction
Nov 20, 2022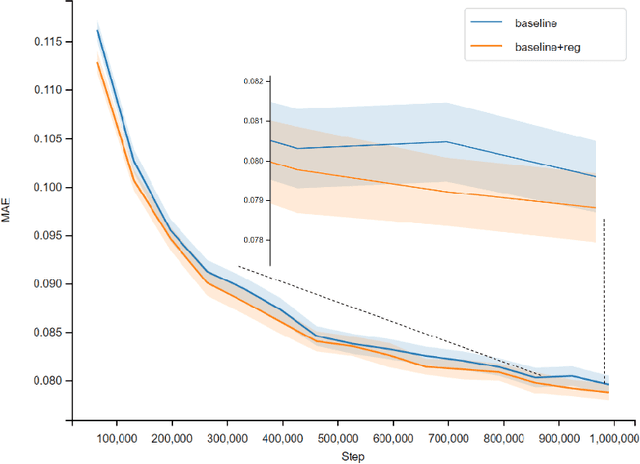
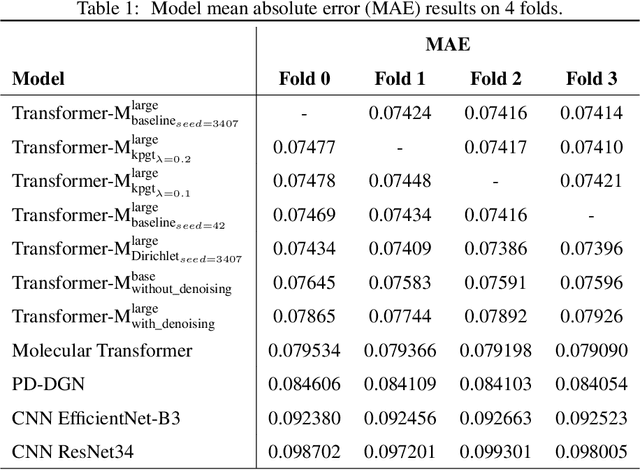
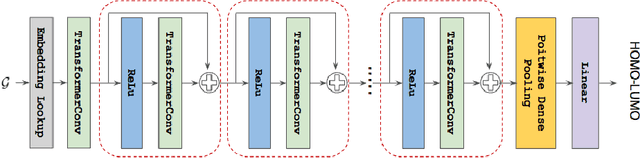
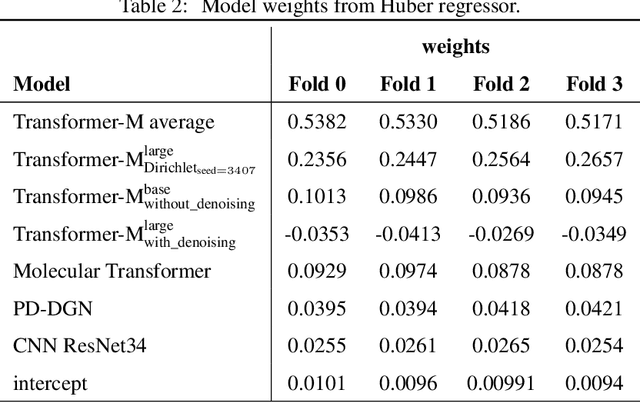
Abstract:Previous works have demonstrated the importance of considering different modalities on molecules, each of which provide a varied granularity of information for downstream property prediction tasks. Our method combines variants of the recent TransformerM architecture with Transformer, GNN, and ResNet backbone architectures. Models are trained on the 2D data, 3D data, and image modalities of molecular graphs. We ensemble these models with a HuberRegressor. The models are trained on 4 different train/validation splits of the original train + valid datasets. This yields a winning solution to the 2\textsuperscript{nd} edition of the OGB Large-Scale Challenge (2022) on the PCQM4Mv2 molecular property prediction dataset. Our proposed method achieves a test-challenge MAE of $0.0723$ and a validation MAE of $0.07145$. Total inference time for our solution is less than 2 hours. We open-source our code at https://github.com/jfpuget/NVIDIA-PCQM4Mv2.
Optimized U-Net for Brain Tumor Segmentation
Oct 07, 2021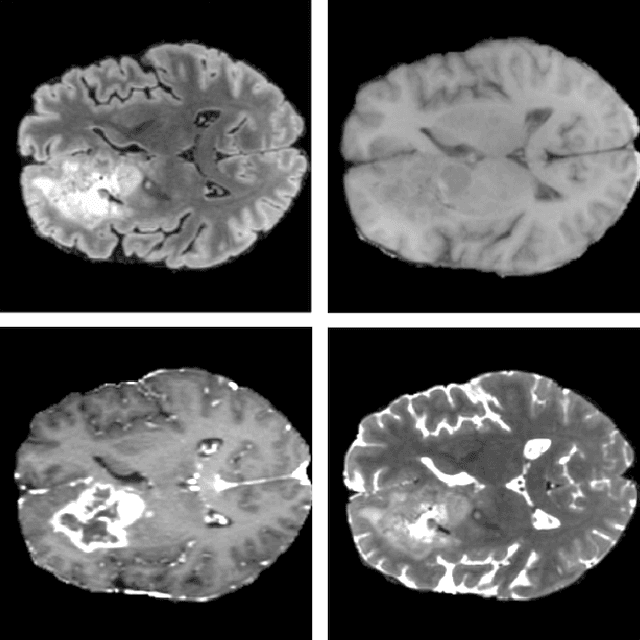
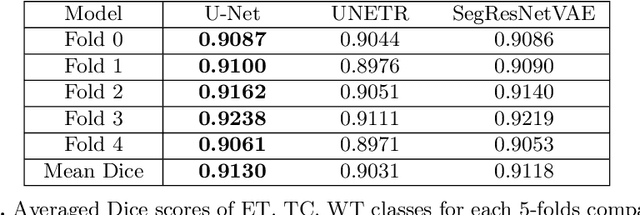
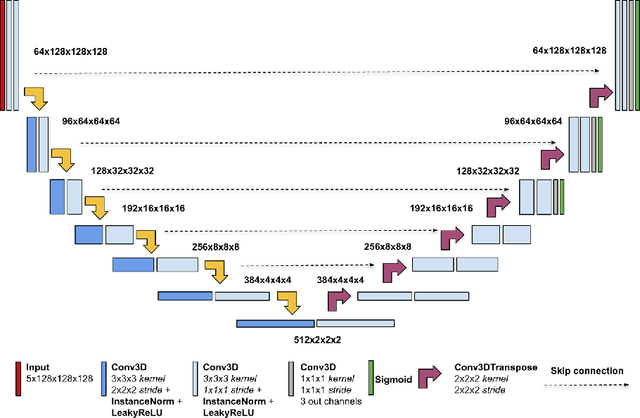
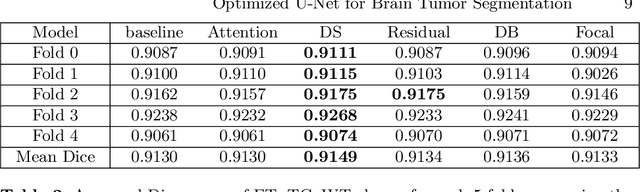
Abstract:We propose an optimized U-Net architecture for a brain \mbox{tumor} segmentation task in the BraTS21 Challenge. To find the \mbox{optimal} model architecture and learning schedule we ran an extensive ablation study to test: deep supervision loss, Focal loss, decoder attention, drop block, and residual connections. Additionally, we have searched for the optimal depth of the U-Net and number of convolutional channels. Our solution was the winner of the challenge validation phase, with the normalized statistical ranking score of 0.267 and mean Dice score of 0.8855
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge