Nils D. Forkert
Exploring the interplay of label bias with subgroup size and separability: A case study in mammographic density classification
Jul 24, 2025Abstract:Systematic mislabelling affecting specific subgroups (i.e., label bias) in medical imaging datasets represents an understudied issue concerning the fairness of medical AI systems. In this work, we investigated how size and separability of subgroups affected by label bias influence the learned features and performance of a deep learning model. Therefore, we trained deep learning models for binary tissue density classification using the EMory BrEast imaging Dataset (EMBED), where label bias affected separable subgroups (based on imaging manufacturer) or non-separable "pseudo-subgroups". We found that simulated subgroup label bias led to prominent shifts in the learned feature representations of the models. Importantly, these shifts within the feature space were dependent on both the relative size and the separability of the subgroup affected by label bias. We also observed notable differences in subgroup performance depending on whether a validation set with clean labels was used to define the classification threshold for the model. For instance, with label bias affecting the majority separable subgroup, the true positive rate for that subgroup fell from 0.898, when the validation set had clean labels, to 0.518, when the validation set had biased labels. Our work represents a key contribution toward understanding the consequences of label bias on subgroup fairness in medical imaging AI.
Towards objective and systematic evaluation of bias in medical imaging AI
Nov 03, 2023



Abstract:Artificial intelligence (AI) models trained using medical images for clinical tasks often exhibit bias in the form of disparities in performance between subgroups. Since not all sources of biases in real-world medical imaging data are easily identifiable, it is challenging to comprehensively assess how those biases are encoded in models, and how capable bias mitigation methods are at ameliorating performance disparities. In this article, we introduce a novel analysis framework for systematically and objectively investigating the impact of biases in medical images on AI models. We developed and tested this framework for conducting controlled in silico trials to assess bias in medical imaging AI using a tool for generating synthetic magnetic resonance images with known disease effects and sources of bias. The feasibility is showcased by using three counterfactual bias scenarios to measure the impact of simulated bias effects on a convolutional neural network (CNN) classifier and the efficacy of three bias mitigation strategies. The analysis revealed that the simulated biases resulted in expected subgroup performance disparities when the CNN was trained on the synthetic datasets. Moreover, reweighing was identified as the most successful bias mitigation strategy for this setup, and we demonstrated how explainable AI methods can aid in investigating the manifestation of bias in the model using this framework. Developing fair AI models is a considerable challenge given that many and often unknown sources of biases can be present in medical imaging datasets. In this work, we present a novel methodology to objectively study the impact of biases and mitigation strategies on deep learning pipelines, which can support the development of clinical AI that is robust and responsible.
Weakly Supervised Medical Image Segmentation With Soft Labels and Noise Robust Loss
Sep 16, 2022
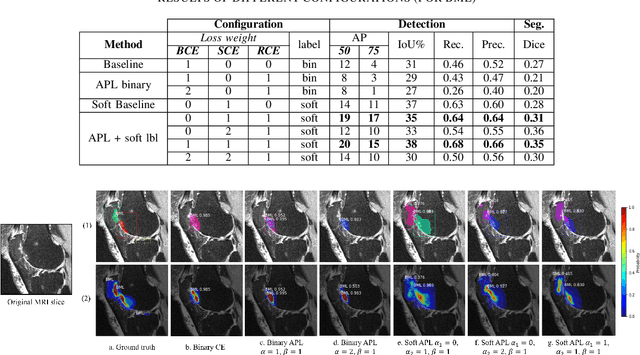
Abstract:Recent advances in deep learning algorithms have led to significant benefits for solving many medical image analysis problems. Training deep learning models commonly requires large datasets with expert-labeled annotations. However, acquiring expert-labeled annotation is not only expensive but also is subjective, error-prone, and inter-/intra- observer variability introduces noise to labels. This is particularly a problem when using deep learning models for segmenting medical images due to the ambiguous anatomical boundaries. Image-based medical diagnosis tools using deep learning models trained with incorrect segmentation labels can lead to false diagnoses and treatment suggestions. Multi-rater annotations might be better suited to train deep learning models with small training sets compared to single-rater annotations. The aim of this paper was to develop and evaluate a method to generate probabilistic labels based on multi-rater annotations and anatomical knowledge of the lesion features in MRI and a method to train segmentation models using probabilistic labels using normalized active-passive loss as a "noise-tolerant loss" function. The model was evaluated by comparing it to binary ground truth for 17 knees MRI scans for clinical segmentation and detection of bone marrow lesions (BML). The proposed method successfully improved precision 14, recall 22, and Dice score 8 percent compared to a binary cross-entropy loss function. Overall, the results of this work suggest that the proposed normalized active-passive loss using soft labels successfully mitigated the effects of noisy labels.
Self-Supervised-RCNN for Medical Image Segmentation with Limited Data Annotation
Jul 17, 2022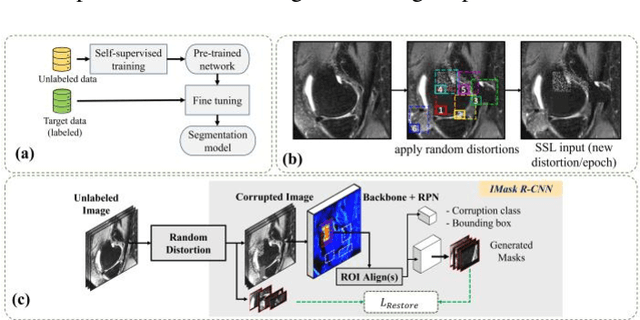
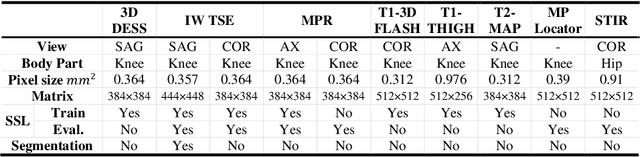
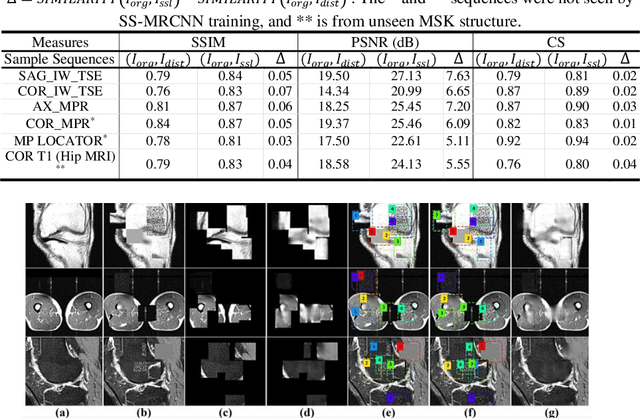
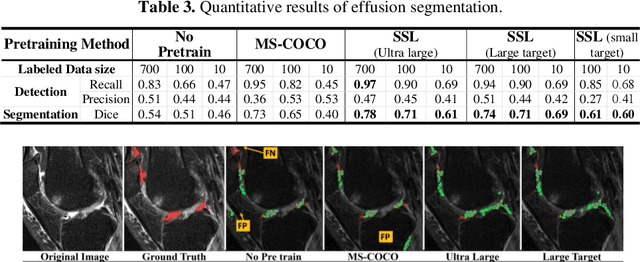
Abstract:Many successful methods developed for medical image analysis that are based on machine learning use supervised learning approaches, which often require large datasets annotated by experts to achieve high accuracy. However, medical data annotation is time-consuming and expensive, especially for segmentation tasks. To solve the problem of learning with limited labeled medical image data, an alternative deep learning training strategy based on self-supervised pretraining on unlabeled MRI scans is proposed in this work. Our pretraining approach first, randomly applies different distortions to random areas of unlabeled images and then predicts the type of distortions and loss of information. To this aim, an improved version of Mask-RCNN architecture has been adapted to localize the distortion location and recover the original image pixels. The effectiveness of the proposed method for segmentation tasks in different pre-training and fine-tuning scenarios is evaluated based on the Osteoarthritis Initiative dataset. Using this self-supervised pretraining method improved the Dice score by 20% compared to training from scratch. The proposed self-supervised learning is simple, effective, and suitable for different ranges of medical image analysis tasks including anomaly detection, segmentation, and classification.
Constructing High-Order Signed Distance Maps from Computed Tomography Data with Application to Bone Morphometry
Nov 02, 2021
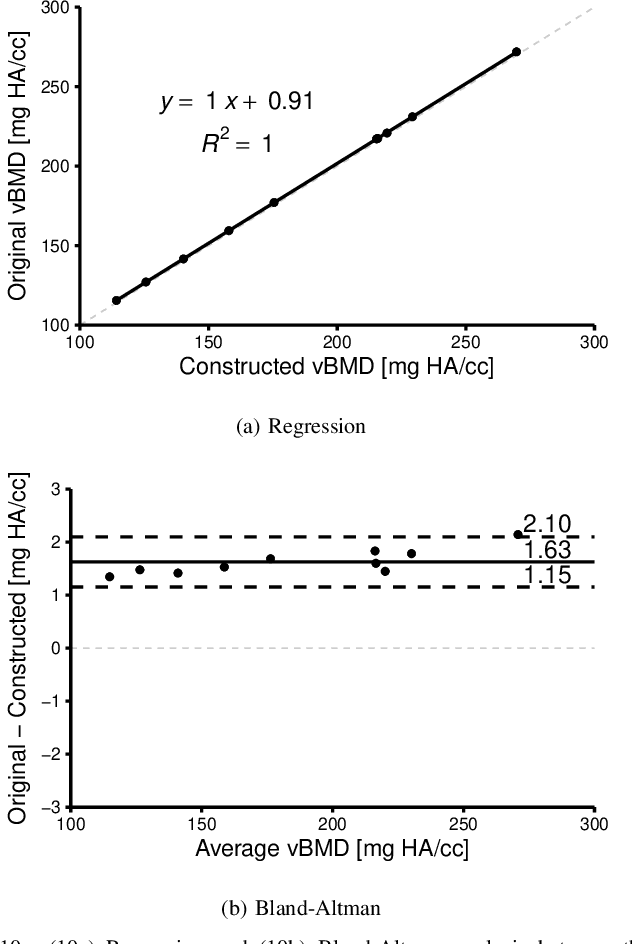

Abstract:An algorithm is presented for constructing high-order signed distance fields for two phase materials imaged with computed tomography. The signed distance field is high-order in that it is free of the quantization artifact associated with the distance transform of sampled signals. The narrowband is solved using a closest point algorithm extended for implicit embeddings that are not a signed distance field. The high-order fast sweeping algorithm is used to extend the narrowband to the remainder of the domain. The order of accuracy of the narrowband and extension methods are verified on ideal implicit surfaces. The method is applied to ten excised cubes of bovine trabecular bone. Localization of the surface, estimation of phase densities, and local morphometry is validated with these subjects. Since the embedding is high-order, gradients and thus curvatures can be accurately estimated locally in the image data.
High-Order Signed Distance Transform of Sampled Signals
Oct 26, 2021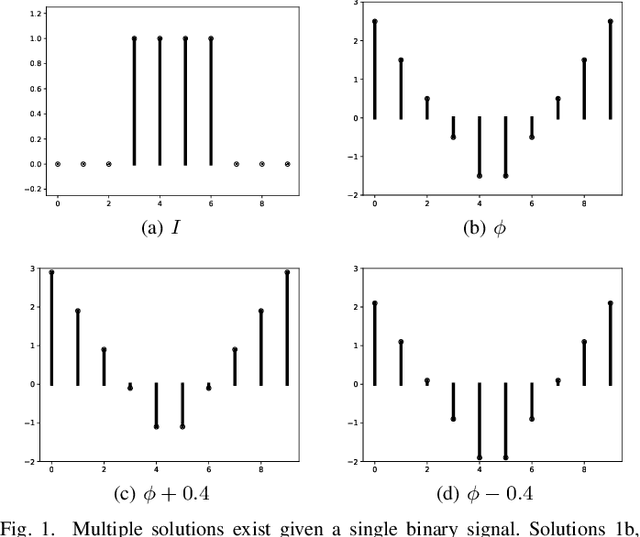
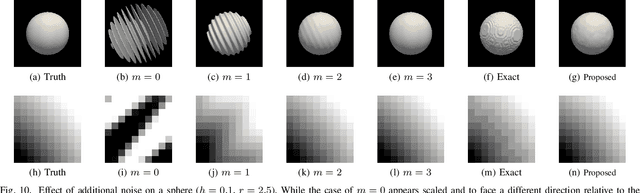
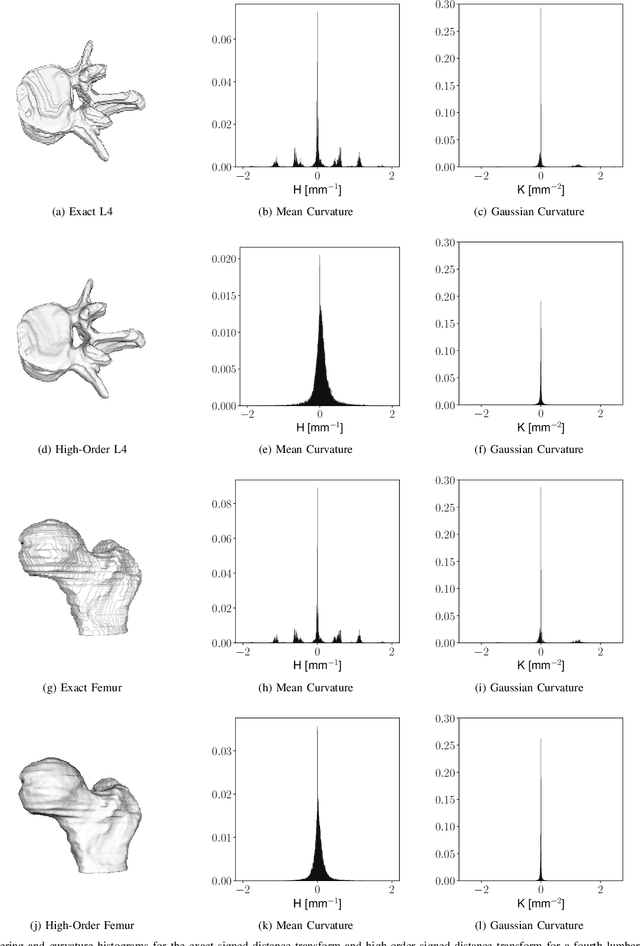
Abstract:Signed distance transforms of sampled signals can be constructed better than the traditional exact signed distance transform. Such a transform is termed the high-order signed distance transform and is defined as satisfying three conditions: the Eikonal equation, recovery by a Heaviside function, and has an order of accuracy greater than unity away from the medial axis. Such a transform is an improvement to the classic notion of an exact signed distance transform because it does not exhibit artifacts of quantization. A large constant, linear time complexity high-order signed distance transform for arbitrary dimensionality sampled signals is developed based on the high order fast sweeping method. The transform is initialized with an exact signed distance transform and quantization corrected through an upwind solver for the boundary value Eikonal equation. The proposed method cannot attain arbitrary order of accuracy and is limited by the initialization method and non-uniqueness of the problem. However, meshed surfaces are visually smoother and do not exhibit artifacts of quantization in local mean and Gaussian curvature.
Local Morphometry of Closed, Implicit Surfaces
Jul 29, 2021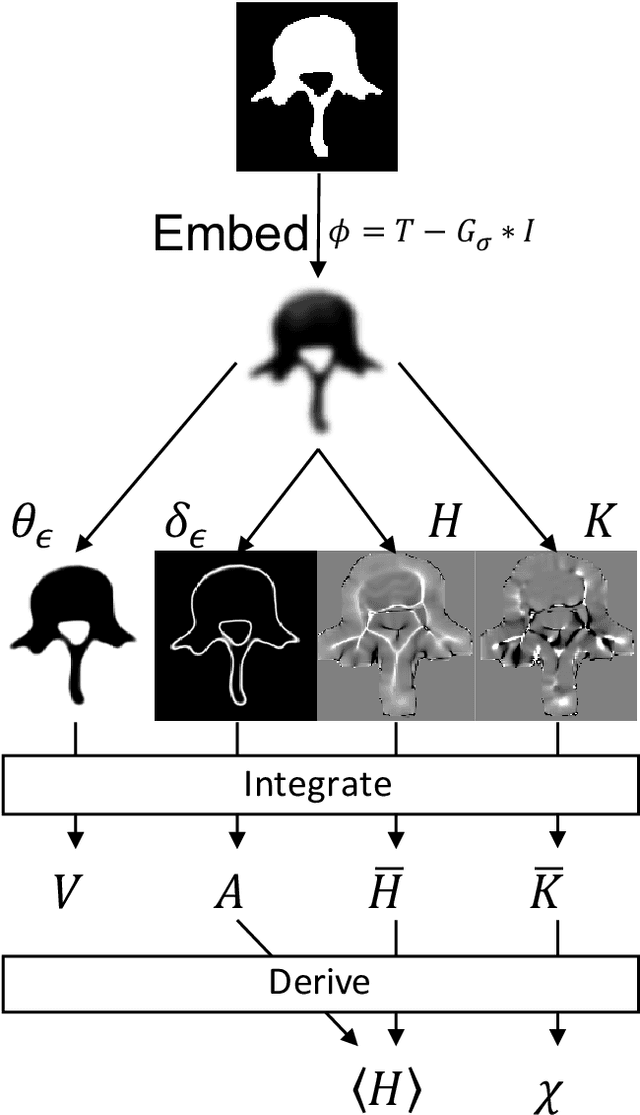
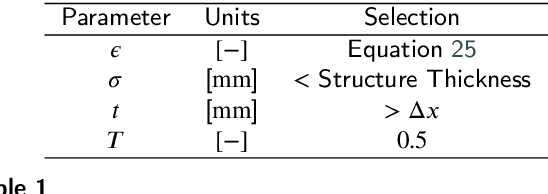
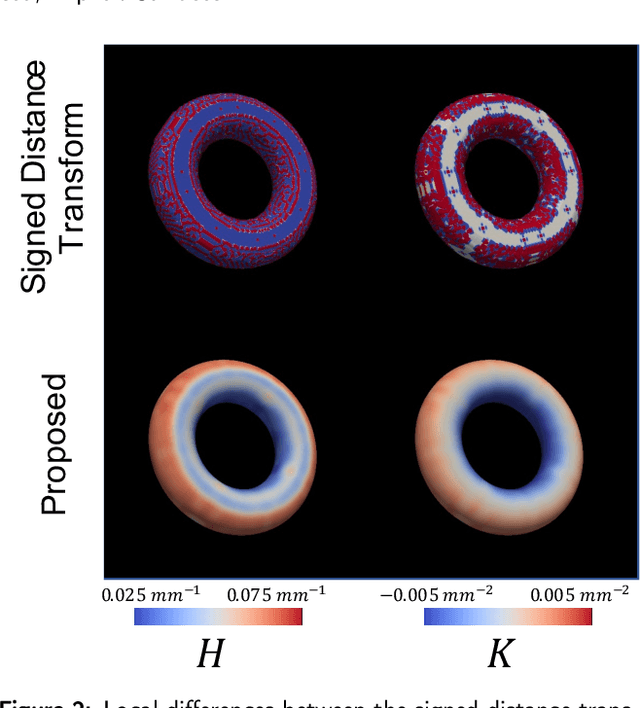
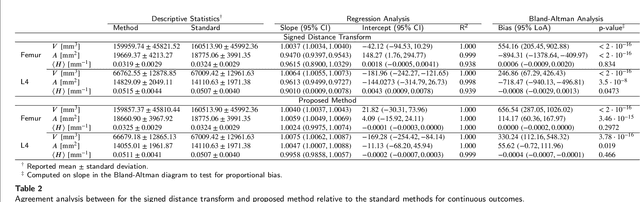
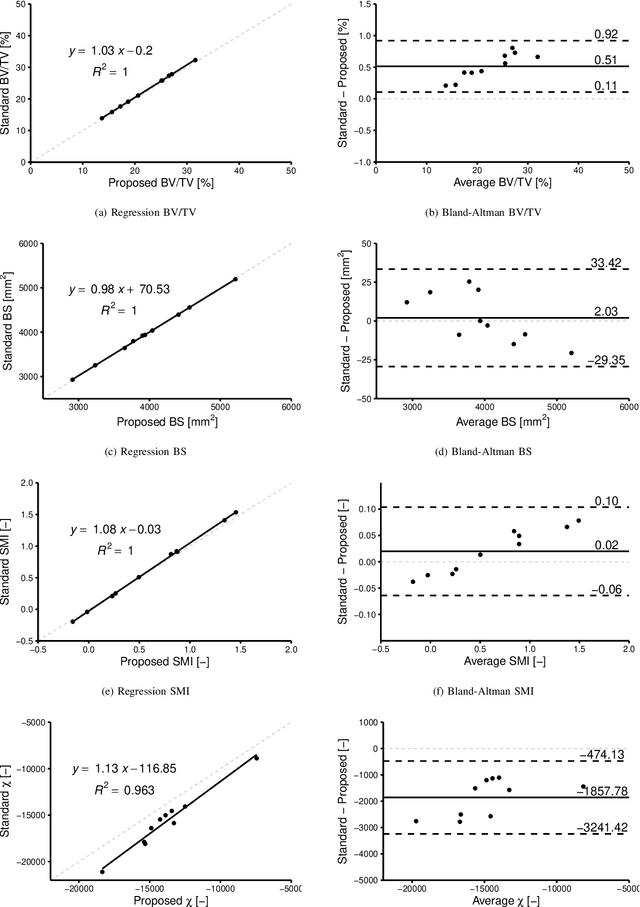
Abstract:Anatomical structures such as the hippocampus, liver, and bones can be analyzed as orientable, closed surfaces. This permits the computation of volume, surface area, mean curvature, Gaussian curvature, and the Euler-Poincar\'e characteristic as well as comparison of these morphometrics between structures of different topology. The structures are commonly represented implicitly in curve evolution problems as the zero level set of an embedding. Practically, binary images of anatomical structures are embedded using a signed distance transform. However, quantization prevents the accurate computation of curvatures, leading to considerable errors in morphometry. This paper presents a fast, simple embedding procedure for accurate local morphometry as the zero crossing of the Gaussian blurred binary image. The proposed method was validated based on the femur and fourth lumbar vertebrae of 50 clinical computed tomography datasets. The results show that the signed distance transform leads to large quantization errors in the computed local curvature. Global validation of morphometry using regression and Bland-Altman analysis revealed that the coefficient of determination for the average mean curvature is improved from 93.8% with the signed distance transform to 100% with the proposed method. For the surface area, the proportional bias is improved from -5.0% for the signed distance transform to +0.6% for the proposed method. The Euler-Poincar\'e characteristic is improved from unusable in the signed distance transform to 98% accuracy for the proposed method. The proposed method enables an improved local and global evaluation of curvature for purposes of morphometry on closed, implicit surfaces.
Bidirectional Modeling and Analysis of Brain Aging with Normalizing Flows
Nov 26, 2020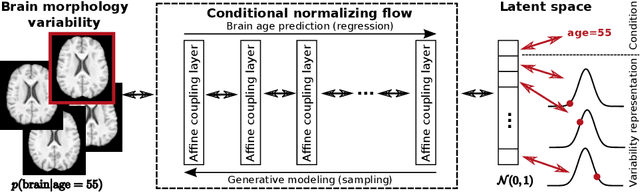
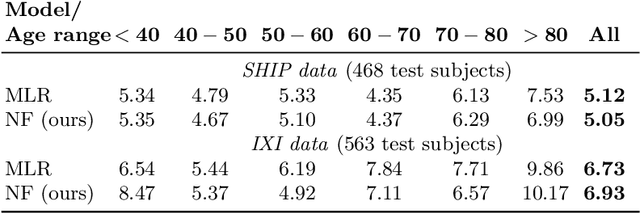
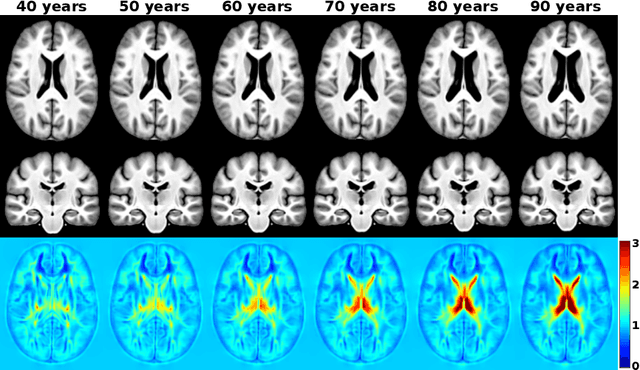
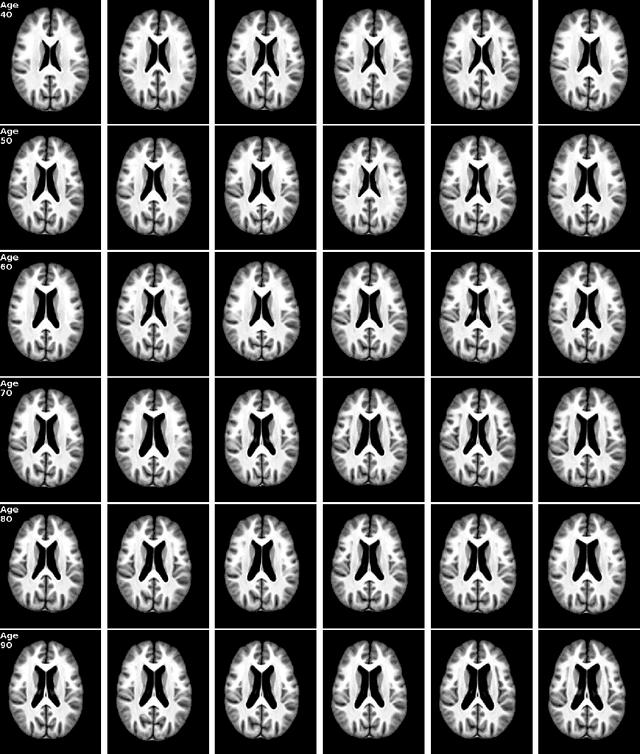
Abstract:Brain aging is a widely studied longitudinal process throughout which the brain undergoes considerable morphological changes and various machine learning approaches have been proposed to analyze it. Within this context, brain age prediction from structural MR images and age-specific brain morphology template generation are two problems that have attracted much attention. While most approaches tackle these tasks independently, we assume that they are inverse directions of the same functional bidirectional relationship between a brain's morphology and an age variable. In this paper, we propose to model this relationship with a single conditional normalizing flow, which unifies brain age prediction and age-conditioned generative modeling in a novel way. In an initial evaluation of this idea, we show that our normalizing flow brain aging model can accurately predict brain age while also being able to generate age-specific brain morphology templates that realistically represent the typical aging trend in a healthy population. This work is a step towards unified modeling of functional relationships between 3D brain morphology and clinical variables of interest with powerful normalizing flows.
DeepVesselNet: Vessel Segmentation, Centerline Prediction, and Bifurcation Detection in 3-D Angiographic Volumes
Mar 25, 2018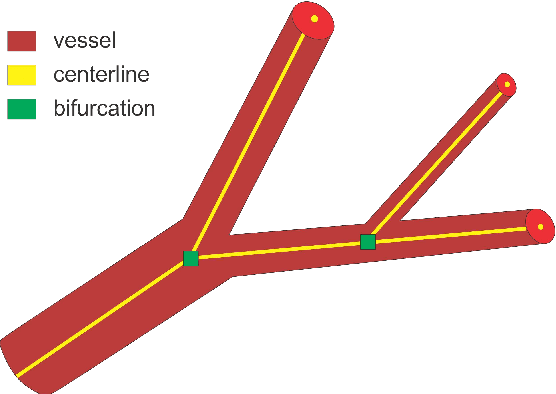


Abstract:We present DeepVesselNet, an architecture tailored to the challenges to be addressed when extracting vessel networks and corresponding features in 3-D angiography using deep learning. We discuss the problems of low execution speed and high memory requirements associated with full 3-D convolutional networks, high class imbalance arising from low percentage (less than 3%) of vessel voxels, and unavailability of accurately annotated training data - and offer solutions that are the building blocks of DeepVesselNet. First, we formulate 2-D orthogonal cross-hair filters which make use of 3-D context information. Second, we introduce a class balancing cross-entropy score with false positive rate correction to handle the high class imbalance and high false positive rate problems associated with existing loss functions. Finally, we generate synthetic dataset using a computational angiogenesis model, capable of generating vascular networks under physiological constraints on local network structure and topology, and use these data for transfer learning. DeepVesselNet is optimized for segmenting vessels, predicting centerlines, and localizing bifurcations. We test the performance on a range of angiographic volumes including clinical Time-of-Flight MRA data of the human brain, as well as synchrotron radiation X-ray tomographic microscopy scans of the rat brain. Our experiments show that, by replacing 3-D filters with 2-D orthogonal cross-hair filters in our network, speed is improved by 23% while accuracy is maintained. Our class balancing metric is crucial for training the network and pre-training with synthetic data helps in early convergence of the training process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge