Self-Supervised-RCNN for Medical Image Segmentation with Limited Data Annotation
Paper and Code
Jul 17, 2022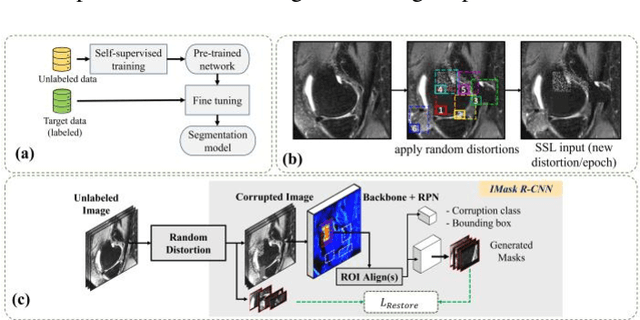
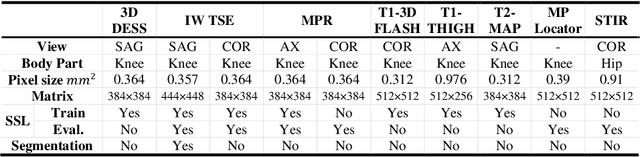
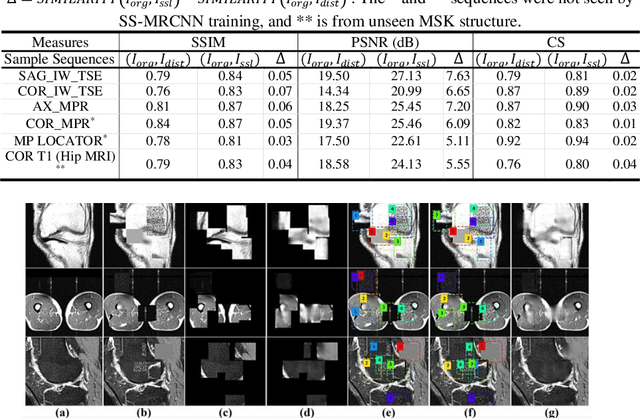
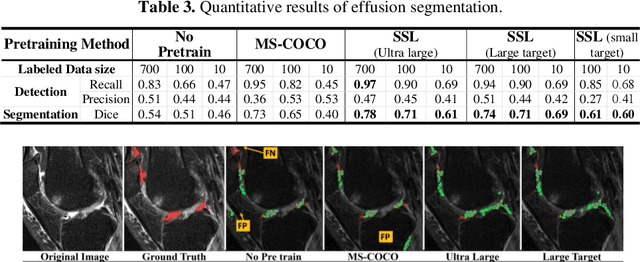
Many successful methods developed for medical image analysis that are based on machine learning use supervised learning approaches, which often require large datasets annotated by experts to achieve high accuracy. However, medical data annotation is time-consuming and expensive, especially for segmentation tasks. To solve the problem of learning with limited labeled medical image data, an alternative deep learning training strategy based on self-supervised pretraining on unlabeled MRI scans is proposed in this work. Our pretraining approach first, randomly applies different distortions to random areas of unlabeled images and then predicts the type of distortions and loss of information. To this aim, an improved version of Mask-RCNN architecture has been adapted to localize the distortion location and recover the original image pixels. The effectiveness of the proposed method for segmentation tasks in different pre-training and fine-tuning scenarios is evaluated based on the Osteoarthritis Initiative dataset. Using this self-supervised pretraining method improved the Dice score by 20% compared to training from scratch. The proposed self-supervised learning is simple, effective, and suitable for different ranges of medical image analysis tasks including anomaly detection, segmentation, and classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge