Pauline Mouches
Automated interictal epileptic spike detection from simple and noisy annotations in MEG data
Oct 24, 2025Abstract:In drug-resistant epilepsy, presurgical evaluation of epilepsy can be considered. Magnetoencephalography (MEG) has been shown to be an effective exam to inform the localization of the epileptogenic zone through the localization of interictal epileptic spikes. Manual detection of these pathological biomarkers remains a fastidious and error-prone task due to the high dimensionality of MEG recordings, and interrater agreement has been reported to be only moderate. Current automated methods are unsuitable for clinical practice, either requiring extensively annotated data or lacking robustness on non-typical data. In this work, we demonstrate that deep learning models can be used for detecting interictal spikes in MEG recordings, even when only temporal and single-expert annotations are available, which represents real-world clinical practice. We propose two model architectures: a feature-based artificial neural network (ANN) and a convolutional neural network (CNN), trained on a database of 59 patients, and evaluated against a state-of-the-art model to classify short time windows of signal. In addition, we employ an interactive machine learning strategy to iteratively improve our data annotation quality using intermediary model outputs. Both proposed models outperform the state-of-the-art model (F1-scores: CNN=0.46, ANN=0.44) when tested on 10 holdout test patients. The interactive machine learning strategy demonstrates that our models are robust to noisy annotations. Overall, results highlight the robustness of models with simple architectures when analyzing complex and imperfectly annotated data. Our method of interactive machine learning offers great potential for faster data annotation, while our models represent useful and efficient tools for automated interictal spikes detection.
Time CNN and Graph Convolution Network for Epileptic Spike Detection in MEG Data
Oct 13, 2023


Abstract:Magnetoencephalography (MEG) recordings of patients with epilepsy exhibit spikes, a typical biomarker of the pathology. Detecting those spikes allows accurate localization of brain regions triggering seizures. Spike detection is often performed manually. However, it is a burdensome and error prone task due to the complexity of MEG data. To address this problem, we propose a 1D temporal convolutional neural network (Time CNN) coupled with a graph convolutional network (GCN) to classify short time frames of MEG recording as containing a spike or not. Compared to other recent approaches, our models have fewer parameters to train and we propose to use a GCN to account for MEG sensors spatial relationships. Our models produce clinically relevant results and outperform deep learning-based state-of-the-art methods reaching a classification f1-score of 76.7% on a balanced dataset and of 25.5% on a realistic, highly imbalanced dataset, for the spike class.
Bidirectional Modeling and Analysis of Brain Aging with Normalizing Flows
Nov 26, 2020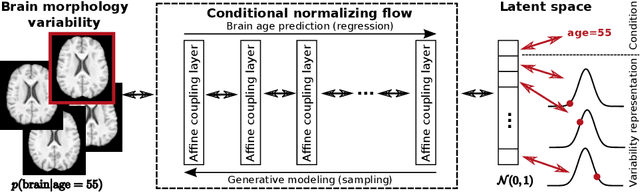
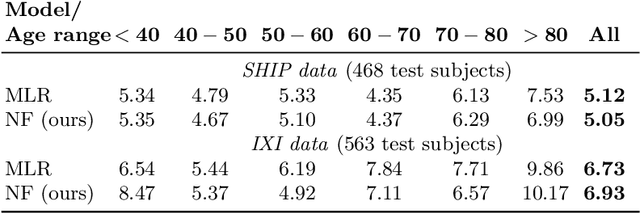
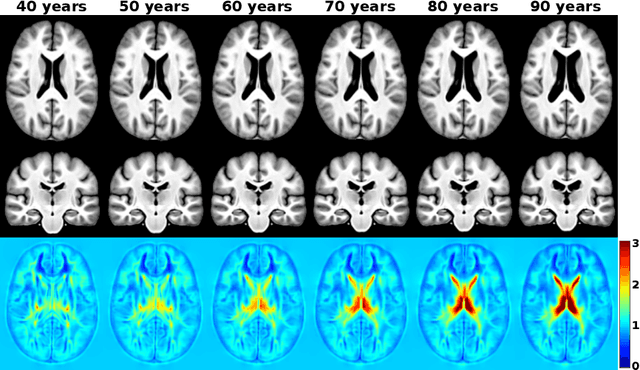
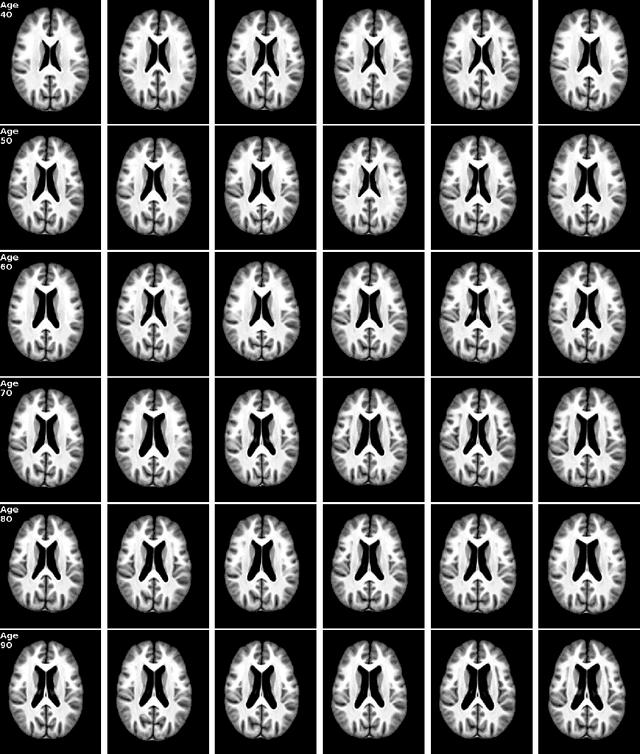
Abstract:Brain aging is a widely studied longitudinal process throughout which the brain undergoes considerable morphological changes and various machine learning approaches have been proposed to analyze it. Within this context, brain age prediction from structural MR images and age-specific brain morphology template generation are two problems that have attracted much attention. While most approaches tackle these tasks independently, we assume that they are inverse directions of the same functional bidirectional relationship between a brain's morphology and an age variable. In this paper, we propose to model this relationship with a single conditional normalizing flow, which unifies brain age prediction and age-conditioned generative modeling in a novel way. In an initial evaluation of this idea, we show that our normalizing flow brain aging model can accurately predict brain age while also being able to generate age-specific brain morphology templates that realistically represent the typical aging trend in a healthy population. This work is a step towards unified modeling of functional relationships between 3D brain morphology and clinical variables of interest with powerful normalizing flows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge