Carole Lartizien
MYRIAD
GAN-based synthetic FDG PET images from T1 brain MRI can serve to improve performance of deep unsupervised anomaly detection models
May 12, 2025Abstract:Background and Objective. Research in the cross-modal medical image translation domain has been very productive over the past few years in tackling the scarce availability of large curated multimodality datasets with the promising performance of GAN-based architectures. However, only a few of these studies assessed task-based related performance of these synthetic data, especially for the training of deep models. Method. We design and compare different GAN-based frameworks for generating synthetic brain [18F]fluorodeoxyglucose (FDG) PET images from T1 weighted MRI data. We first perform standard qualitative and quantitative visual quality evaluation. Then, we explore further impact of using these fake PET data in the training of a deep unsupervised anomaly detection (UAD) model designed to detect subtle epilepsy lesions in T1 MRI and FDG PET images. We introduce novel diagnostic task-oriented quality metrics of the synthetic FDG PET data tailored to our unsupervised detection task, then use these fake data to train a use case UAD model combining a deep representation learning based on siamese autoencoders with a OC-SVM density support estimation model. This model is trained on normal subjects only and allows the detection of any variation from the pattern of the normal population. We compare the detection performance of models trained on 35 paired real MR T1 of normal subjects paired either on 35 true PET images or on 35 synthetic PET images generated from the best performing generative models. Performance analysis is conducted on 17 exams of epilepsy patients undergoing surgery. Results. The best performing GAN-based models allow generating realistic fake PET images of control subject with SSIM and PSNR values around 0.9 and 23.8, respectively and in distribution (ID) with regard to the true control dataset. The best UAD model trained on these synthetic normative PET data allows reaching 74% sensitivity. Conclusion. Our results confirm that GAN-based models are the best suited for MR T1 to FDG PET translation, outperforming transformer or diffusion models. We also demonstrate the diagnostic value of these synthetic data for the training of UAD models and evaluation on clinical exams of epilepsy patients. Our code and the normative image dataset are available.
Weakly supervised deep learning model with size constraint for prostate cancer detection in multiparametric MRI and generalization to unseen domains
Nov 04, 2024Abstract:Fully supervised deep models have shown promising performance for many medical segmentation tasks. Still, the deployment of these tools in clinics is limited by the very timeconsuming collection of manually expert-annotated data. Moreover, most of the state-ofthe-art models have been trained and validated on moderately homogeneous datasets. It is known that deep learning methods are often greatly degraded by domain or label shifts and are yet to be built in such a way as to be robust to unseen data or label distributions. In the clinical setting, this problematic is particularly relevant as the deployment institutions may have different scanners or acquisition protocols than those from which the data has been collected to train the model. In this work, we propose to address these two challenges on the detection of clinically significant prostate cancer (csPCa) from bi-parametric MRI. We evaluate the method proposed by (Kervadec et al., 2018), which introduces a size constaint loss to produce fine semantic cancer lesions segmentations from weak circle scribbles annotations. Performance of the model is based on two public (PI-CAI and Prostate158) and one private databases. First, we show that the model achieves on-par performance with strong fully supervised baseline models, both on in-distribution validation data and unseen test images. Second, we observe a performance decrease for both fully supervised and weakly supervised models when tested on unseen data domains. This confirms the crucial need for efficient domain adaptation methods if deep learning models are aimed to be deployed in a clinical environment. Finally, we show that ensemble predictions from multiple trainings increase generalization performance.
Whole-brain radiomics for clustered federated personalization in brain tumor segmentation
Oct 17, 2023



Abstract:Federated learning and its application to medical image segmentation have recently become a popular research topic. This training paradigm suffers from statistical heterogeneity between participating institutions' local datasets, incurring convergence slowdown as well as potential accuracy loss compared to classical training. To mitigate this effect, federated personalization emerged as the federated optimization of one model per institution. We propose a novel personalization algorithm tailored to the feature shift induced by the usage of different scanners and acquisition parameters by different institutions. This method is the first to account for both inter and intra-institution feature shift (multiple scanners used in a single institution). It is based on the computation, within each centre, of a series of radiomic features capturing the global texture of each 3D image volume, followed by a clustering analysis pooling all feature vectors transferred from the local institutions to the central server. Each computed clustered decentralized dataset (potentially including data from different institutions) then serves to finetune a global model obtained through classical federated learning. We validate our approach on the Federated Brain Tumor Segmentation 2022 Challenge dataset (FeTS2022). Our code is available at (https://github.com/MatthisManthe/radiomics_CFFL).
Time CNN and Graph Convolution Network for Epileptic Spike Detection in MEG Data
Oct 13, 2023


Abstract:Magnetoencephalography (MEG) recordings of patients with epilepsy exhibit spikes, a typical biomarker of the pathology. Detecting those spikes allows accurate localization of brain regions triggering seizures. Spike detection is often performed manually. However, it is a burdensome and error prone task due to the complexity of MEG data. To address this problem, we propose a 1D temporal convolutional neural network (Time CNN) coupled with a graph convolutional network (GCN) to classify short time frames of MEG recording as containing a spike or not. Compared to other recent approaches, our models have fewer parameters to train and we propose to use a GCN to account for MEG sensors spatial relationships. Our models produce clinically relevant results and outperform deep learning-based state-of-the-art methods reaching a classification f1-score of 76.7% on a balanced dataset and of 25.5% on a realistic, highly imbalanced dataset, for the spike class.
Towards frugal unsupervised detection of subtle abnormalities in medical imaging
Sep 04, 2023Abstract:Anomaly detection in medical imaging is a challenging task in contexts where abnormalities are not annotated. This problem can be addressed through unsupervised anomaly detection (UAD) methods, which identify features that do not match with a reference model of normal profiles. Artificial neural networks have been extensively used for UAD but they do not generally achieve an optimal trade-o$\hookleftarrow$ between accuracy and computational demand. As an alternative, we investigate mixtures of probability distributions whose versatility has been widely recognized for a variety of data and tasks, while not requiring excessive design e$\hookleftarrow$ort or tuning. Their expressivity makes them good candidates to account for complex multivariate reference models. Their much smaller number of parameters makes them more amenable to interpretation and e cient learning. However, standard estimation procedures, such as the Expectation-Maximization algorithm, do not scale well to large data volumes as they require high memory usage. To address this issue, we propose to incrementally compute inferential quantities. This online approach is illustrated on the challenging detection of subtle abnormalities in MR brain scans for the follow-up of newly diagnosed Parkinsonian patients. The identified structural abnormalities are consistent with the disease progression, as accounted by the Hoehn and Yahr scale.
Anomaly detection in image or latent space of patch-based auto-encoders for industrial image analysis
Jul 04, 2023Abstract:We study several methods for detecting anomalies in color images, constructed on patch-based auto-encoders. Wecompare the performance of three types of methods based, first, on the error between the original image and its reconstruction,second, on the support estimation of the normal image distribution in the latent space, and third, on the error between the originalimage and a restored version of the reconstructed image. These methods are evaluated on the industrial image database MVTecADand compared to two competitive state-of-the-art methods.
One-Class SVM on siamese neural network latent space for Unsupervised Anomaly Detection on brain MRI White Matter Hyperintensities
Apr 17, 2023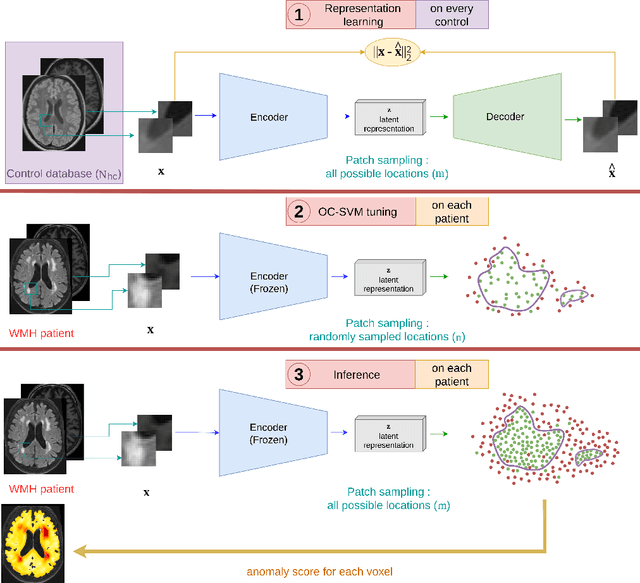
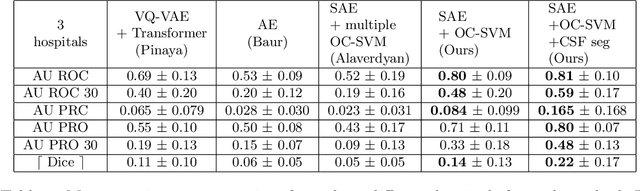
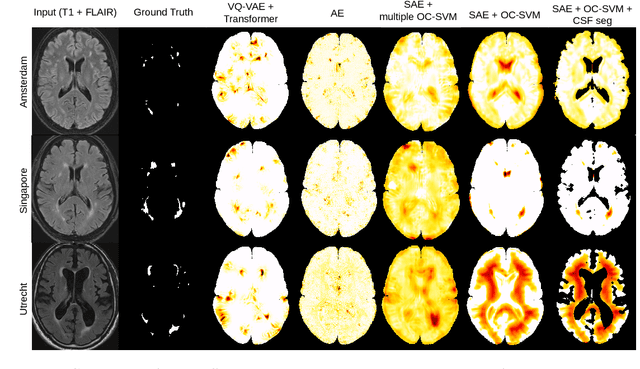
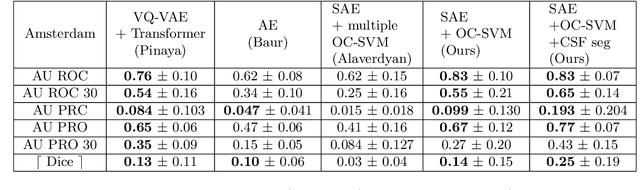
Abstract:Anomaly detection remains a challenging task in neuroimaging when little to no supervision is available and when lesions can be very small or with subtle contrast. Patch-based representation learning has shown powerful representation capacities when applied to industrial or medical imaging and outlier detection methods have been applied successfully to these images. In this work, we propose an unsupervised anomaly detection (UAD) method based on a latent space constructed by a siamese patch-based auto-encoder and perform the outlier detection with a One-Class SVM training paradigm tailored to the lesion detection task in multi-modality neuroimaging. We evaluate performances of this model on a public database, the White Matter Hyperintensities (WMH) challenge and show in par performance with the two best performing state-of-the-art methods reported so far.
Brain subtle anomaly detection based on auto-encoders latent space analysis : application to de novo parkinson patients
Feb 27, 2023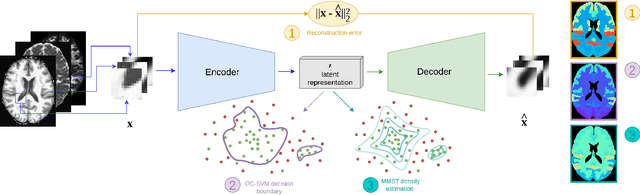
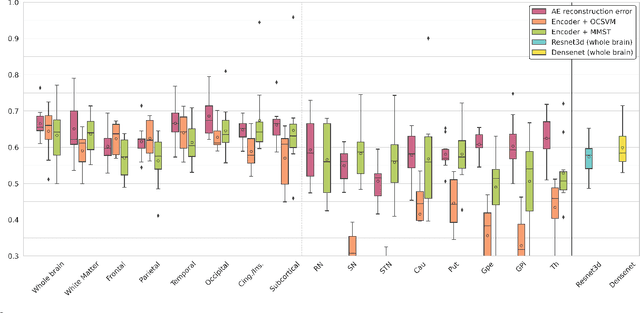
Abstract:Neural network-based anomaly detection remains challenging in clinical applications with little or no supervised information and subtle anomalies such as hardly visible brain lesions. Among unsupervised methods, patch-based auto-encoders with their efficient representation power provided by their latent space, have shown good results for visible lesion detection. However, the commonly used reconstruction error criterion may limit their performance when facing less obvious lesions. In this work, we design two alternative detection criteria. They are derived from multivariate analysis and can more directly capture information from latent space representations. Their performance compares favorably with two additional supervised learning methods, on a difficult de novo Parkinson Disease (PD) classification task.
ProstAttention-Net: A deep attention model for prostate cancer segmentation by aggressiveness in MRI scans
Nov 23, 2022



Abstract:Multiparametric magnetic resonance imaging (mp-MRI) has shown excellent results in the detection of prostate cancer (PCa). However, characterizing prostate lesions aggressiveness in mp-MRI sequences is impossible in clinical practice, and biopsy remains the reference to determine the Gleason score (GS). In this work, we propose a novel end-to-end multi-class network that jointly segments the prostate gland and cancer lesions with GS group grading. After encoding the information on a latent space, the network is separated in two branches: 1) the first branch performs prostate segmentation 2) the second branch uses this zonal prior as an attention gate for the detection and grading of prostate lesions. The model was trained and validated with a 5-fold cross-validation on an heterogeneous series of 219 MRI exams acquired on three different scanners prior prostatectomy. In the free-response receiver operating characteristics (FROC) analysis for clinically significant lesions (defined as GS > 6) detection, our model achieves 69.0% $\pm$14.5% sensitivity at 2.9 false positive per patient on the whole prostate and 70.8% $\pm$14.4% sensitivity at 1.5 false positive when considering the peripheral zone (PZ) only. Regarding the automatic GS group
Perfusion imaging in deep prostate cancer detection from mp-MRI: can we take advantage of it?
Jul 06, 2022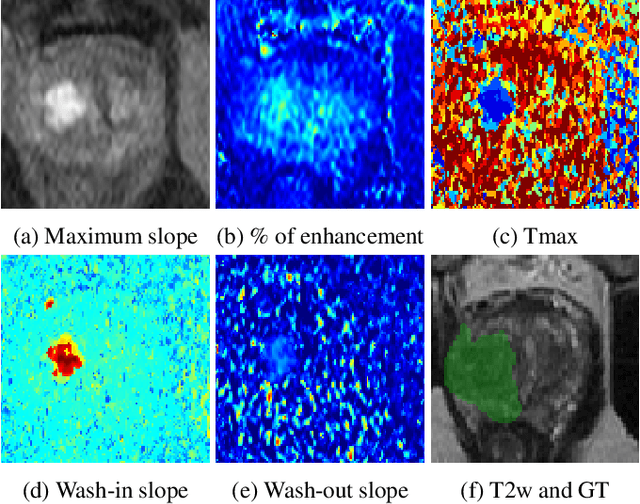

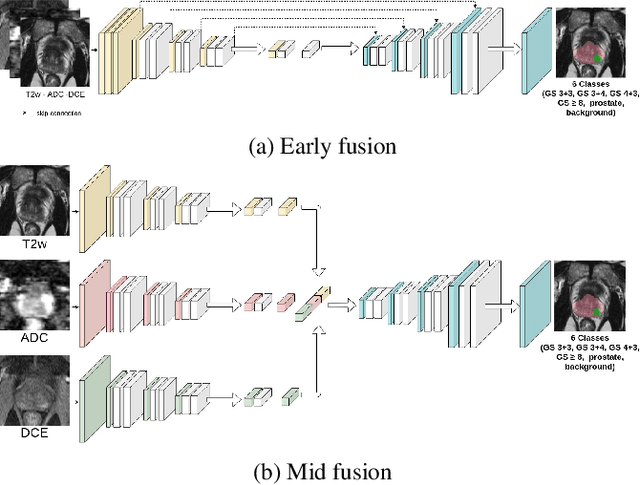

Abstract:To our knowledge, all deep computer-aided detection and diagnosis (CAD) systems for prostate cancer (PCa) detection consider bi-parametric magnetic resonance imaging (bp-MRI) only, including T2w and ADC sequences while excluding the 4D perfusion sequence,which is however part of standard clinical protocols for this diagnostic task. In this paper, we question strategies to integrate information from perfusion imaging in deep neural architectures. To do so, we evaluate several ways to encode the perfusion information in a U-Net like architecture, also considering early versus mid fusion strategies. We compare performance of multiparametric MRI (mp-MRI) models with the baseline bp-MRI model based on a private dataset of 219 mp-MRI exams. Perfusion maps derived from dynamic contrast enhanced MR exams are shown to positively impact segmentation and grading performance of PCa lesions, especially the 3D MR volume corresponding to the maximum slope of the wash-in curve as well as Tmax perfusion maps. The latter mp-MRI models indeed outperform the bp-MRI one whatever the fusion strategy, with Cohen's kappa score of 0.318$\pm$0.019 for the bp-MRI model and 0.378 $\pm$ 0.033 for the model including the maximum slope with a mid fusion strategy, also achieving competitive Cohen's kappa score compared to state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge