Nicola Ferrier
Adversarial Predictions of Data Distributions Across Federated Internet-of-Things Devices
Aug 28, 2023Abstract:Federated learning (FL) is increasingly becoming the default approach for training machine learning models across decentralized Internet-of-Things (IoT) devices. A key advantage of FL is that no raw data are communicated across the network, providing an immediate layer of privacy. Despite this, recent works have demonstrated that data reconstruction can be done with the locally trained model updates which are communicated across the network. However, many of these works have limitations with regard to how the gradients are computed in backpropagation. In this work, we demonstrate that the model weights shared in FL can expose revealing information about the local data distributions of IoT devices. This leakage could expose sensitive information to malicious actors in a distributed system. We further discuss results which show that injecting noise into model weights is ineffective at preventing data leakage without seriously harming the global model accuracy.
EXSCLAIM! -- An automated pipeline for the construction of labeled materials imaging datasets from literature
Mar 19, 2021



Abstract:Due to recent improvements in image resolution and acquisition speed, materials microscopy is experiencing an explosion of published imaging data. The standard publication format, while sufficient for traditional data ingestion scenarios where a select number of images can be critically examined and curated manually, is not conducive to large-scale data aggregation or analysis, hindering data sharing and reuse. Most images in publications are presented as components of a larger figure with their explicit context buried in the main body or caption text, so even if aggregated, collections of images with weak or no digitized contextual labels have limited value. To solve the problem of curating labeled microscopy data from literature, this work introduces the EXSCLAIM! Python toolkit for the automatic EXtraction, Separation, and Caption-based natural Language Annotation of IMages from scientific literature. We highlight the methodology behind the construction of EXSCLAIM! and demonstrate its ability to extract and label open-source scientific images at high volume.
A Two-stage Framework for Compound Figure Separation
Jan 25, 2021



Abstract:Scientific literature contains large volumes of complex, unstructured figures that are compound in nature (i.e. composed of multiple images, graphs, and drawings). Separation of these compound figures is critical for information retrieval from these figures. In this paper, we propose a new strategy for compound figure separation, which decomposes the compound figures into constituent subfigures while preserving the association between the subfigures and their respective caption components. We propose a two-stage framework to address the proposed compound figure separation problem. In particular, the subfigure label detection module detects all subfigure labels in the first stage. Then, in the subfigure detection module, the detected subfigure labels help to detect the subfigures by optimizing the feature selection process and providing the global layout information as extra features. Extensive experiments are conducted to validate the effectiveness and superiority of the proposed framework, which improves the detection precision by 9%.
Scaling Distributed Training of Flood-Filling Networks on HPC Infrastructure for Brain Mapping
May 13, 2019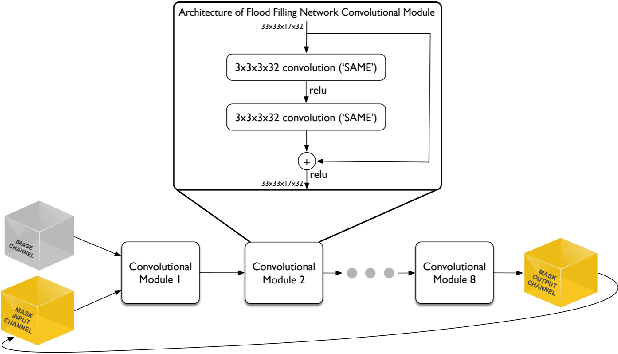
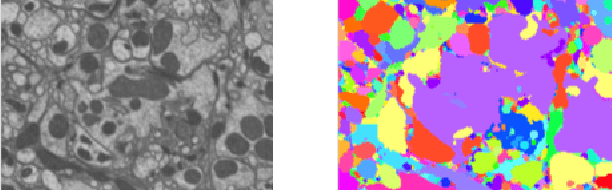
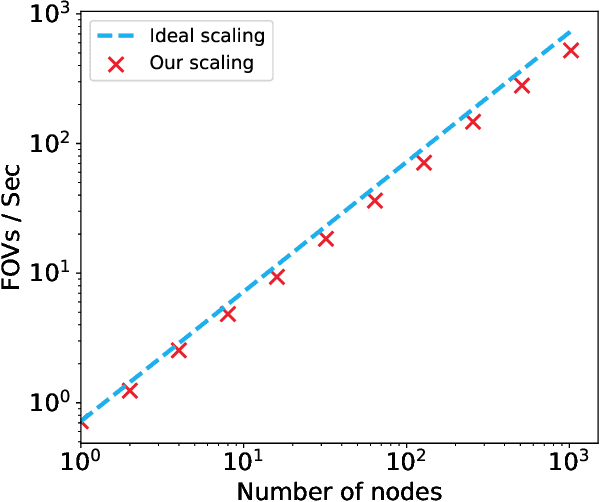
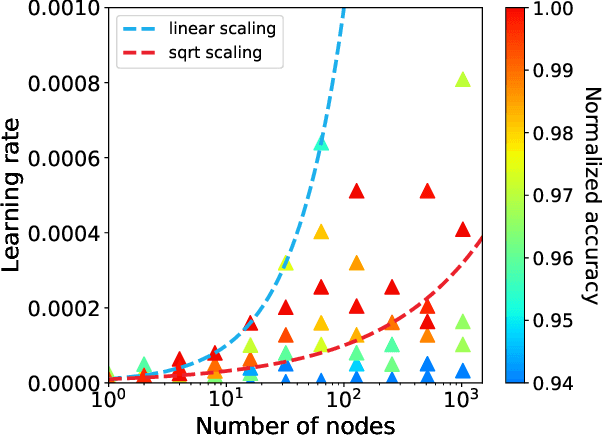
Abstract:Mapping all the neurons in the brain requires automatic reconstruction of entire cells from volume electron microscopy data. The flood-filling networks (FFN) architecture can achieve leading performance. However, the training of the network is computationally very expensive. In order to reduce the training time, we implemented synchronous and data-parallel distributed training using the Horovod framework on top of the published FFN code. We demonstrated the scaling of FFN training up to 1024 Intel Knights Landing (KNL) nodes at Argonne Leadership Computing Facility. We investigated the training accuracy with different optimizers, learning rates, and optional warm-up periods. We discovered that square root scaling for learning rate works best beyond 16 nodes, which is contrary to the case of smaller number of nodes, where linear learning rate scaling with warm-up performs the best. Our distributed training reaches 95% accuracy in approximately 4.5 hours on 1024 KNL nodes using Adam optimizer.
U-SLADS: Unsupervised Learning Approach for Dynamic Dendrite Sampling
Jul 06, 2018
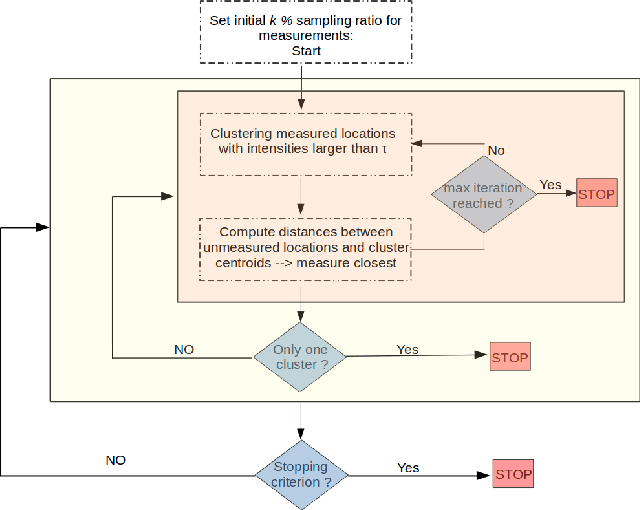
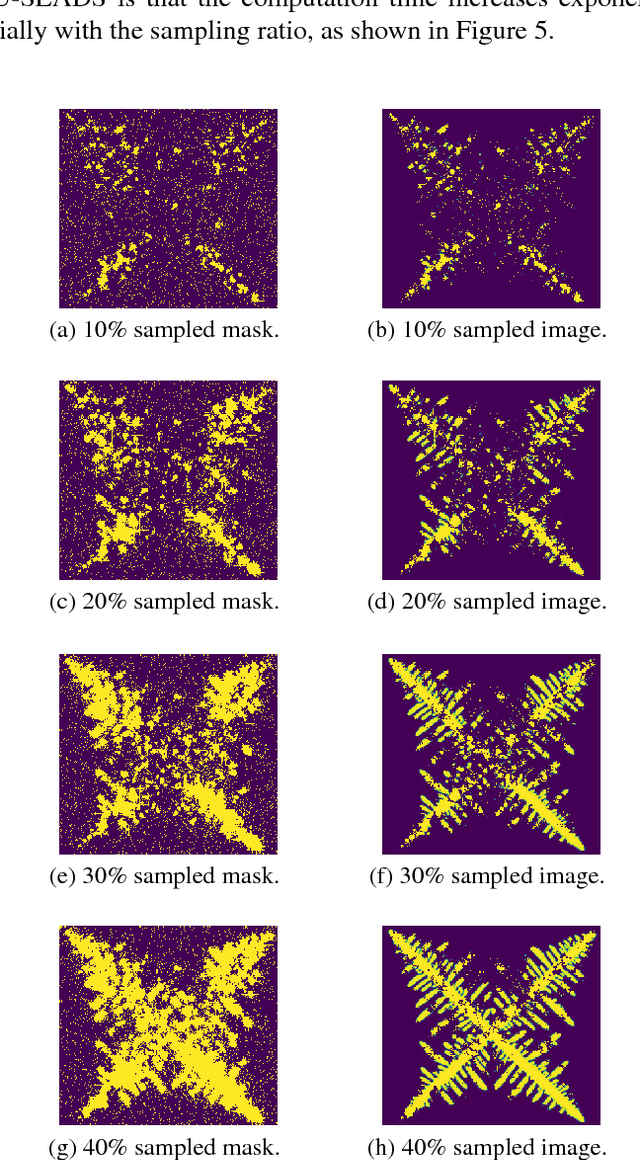
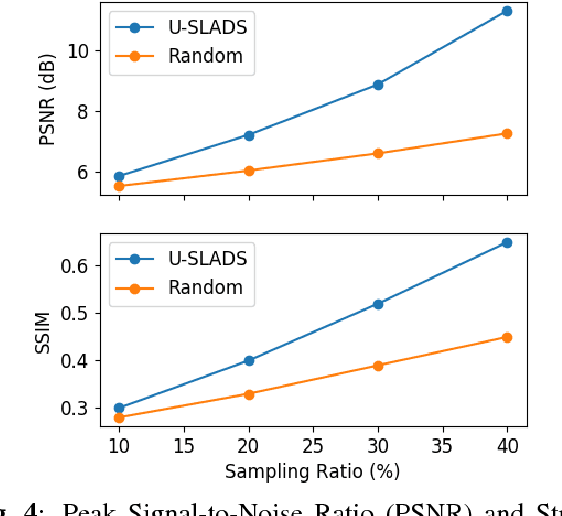
Abstract:Novel data acquisition schemes have been an emerging need for scanning microscopy based imaging techniques to reduce the time in data acquisition and to minimize probing radiation in sample exposure. Varies sparse sampling schemes have been studied and are ideally suited for such applications where the images can be reconstructed from a sparse set of measurements. Dynamic sparse sampling methods, particularly supervised learning based iterative sampling algorithms, have shown promising results for sampling pixel locations on the edges or boundaries during imaging. However, dynamic sampling for imaging skeleton-like objects such as metal dendrites remains difficult. Here, we address a new unsupervised learning approach using Hierarchical Gaussian Mixture Mod- els (HGMM) to dynamically sample metal dendrites. This technique is very useful if the users are interested in fast imaging the primary and secondary arms of metal dendrites in solidification process in materials science.
Reduced Electron Exposure for Energy-Dispersive Spectroscopy using Dynamic Sampling
Jun 27, 2017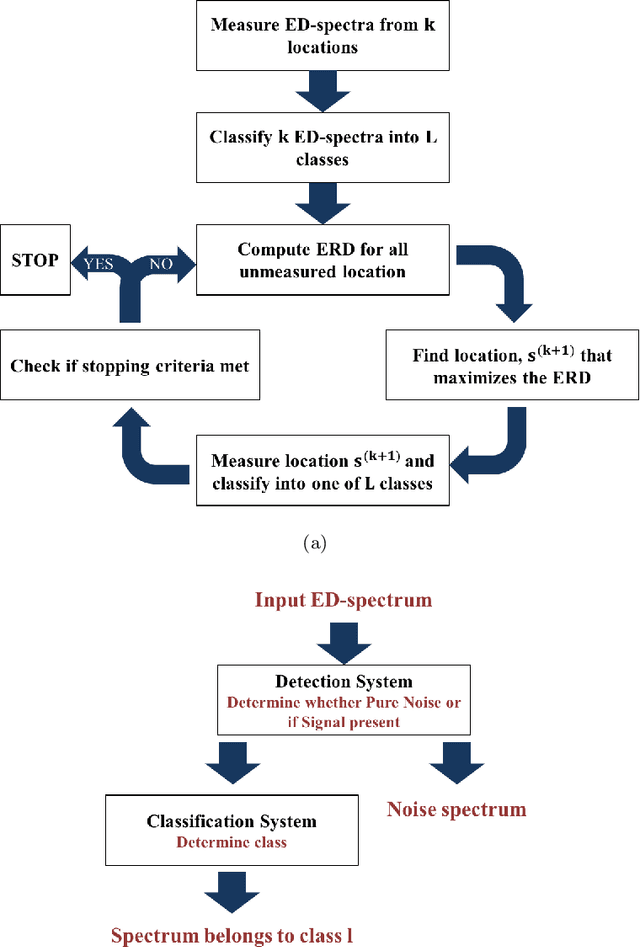
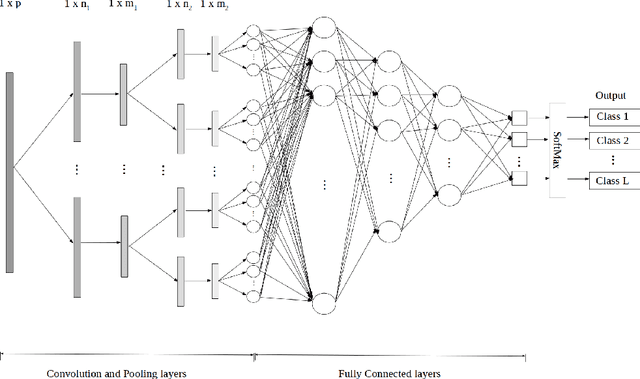
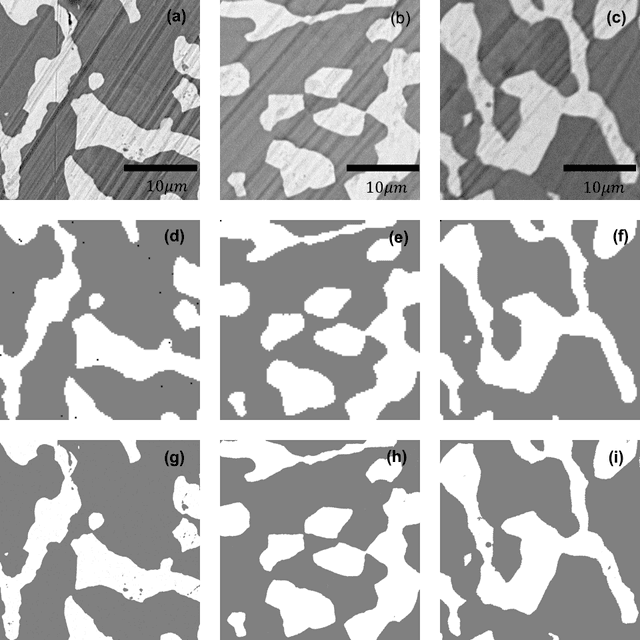
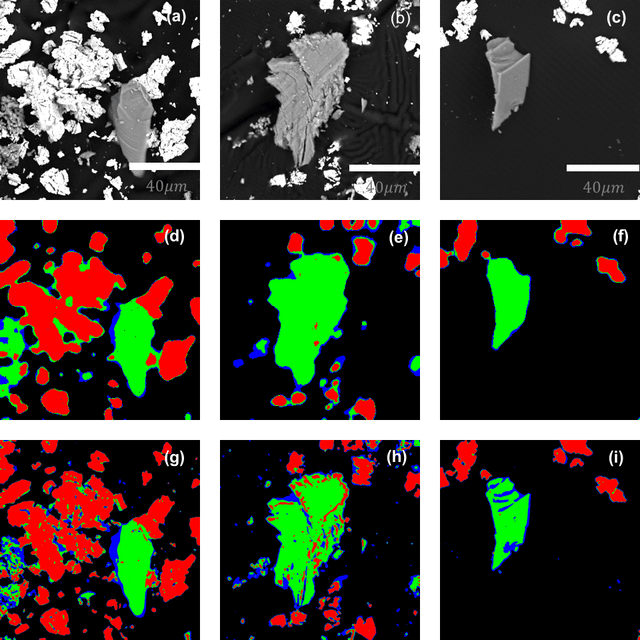
Abstract:Analytical electron microscopy and spectroscopy of biological specimens, polymers, and other beam sensitive materials has been a challenging area due to irradiation damage. There is a pressing need to develop novel imaging and spectroscopic imaging methods that will minimize such sample damage as well as reduce the data acquisition time. The latter is useful for high-throughput analysis of materials structure and chemistry. In this work, we present a novel machine learning based method for dynamic sparse sampling of EDS data using a scanning electron microscope. Our method, based on the supervised learning approach for dynamic sampling algorithm and neural networks based classification of EDS data, allows a dramatic reduction in the total sampling of up to 90%, while maintaining the fidelity of the reconstructed elemental maps and spectroscopic data. We believe this approach will enable imaging and elemental mapping of materials that would otherwise be inaccessible to these analysis techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge