Maria K. Y. Chan
Machine learning for interpreting coherent X-ray speckle patterns
Nov 15, 2022Abstract:Speckle patterns produced by coherent X-ray have a close relationship with the internal structure of materials but quantitative inversion of the relationship to determine structure from images is challenging. Here, we investigate the link between coherent X-ray speckle patterns and sample structures using a model 2D disk system and explore the ability of machine learning to learn aspects of the relationship. Specifically, we train a deep neural network to classify the coherent X-ray speckle pattern images according to the disk number density in the corresponding structure. It is demonstrated that the classification system is accurate for both non-disperse and disperse size distributions.
Ensemble of Pre-Trained Neural Networks for Segmentation and Quality Detection of Transmission Electron Microscopy Images
Sep 05, 2022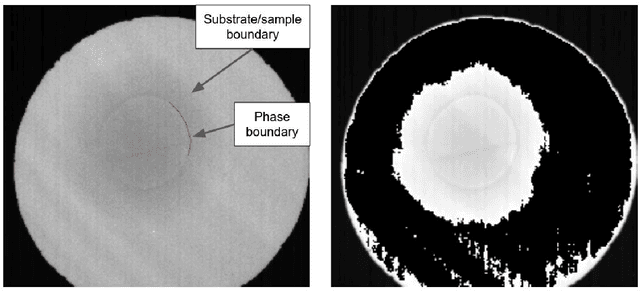

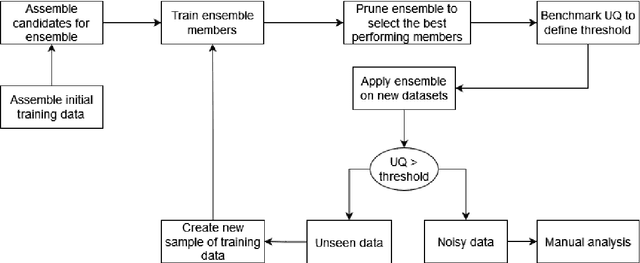
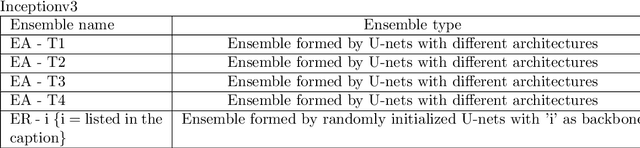
Abstract:Automated analysis of electron microscopy datasets poses multiple challenges, such as limitation in the size of the training dataset, variation in data distribution induced by variation in sample quality and experiment conditions, etc. It is crucial for the trained model to continue to provide acceptable segmentation/classification performance on new data, and quantify the uncertainty associated with its predictions. Among the broad applications of machine learning, various approaches have been adopted to quantify uncertainty, such as Bayesian modeling, Monte Carlo dropout, ensembles, etc. With the aim of addressing the challenges specific to the data domain of electron microscopy, two different types of ensembles of pre-trained neural networks were implemented in this work. The ensembles performed semantic segmentation of ice crystal within a two-phase mixture, thereby tracking its phase transformation to water. The first ensemble (EA) is composed of U-net style networks having different underlying architectures, whereas the second series of ensembles (ER-i) are composed of randomly initialized U-net style networks, wherein each base learner has the same underlying architecture 'i'. The encoders of the base learners were pre-trained on the Imagenet dataset. The performance of EA and ER were evaluated on three different metrics: accuracy, calibration, and uncertainty. It is seen that EA exhibits a greater classification accuracy and is better calibrated, as compared to ER. While the uncertainty quantification of these two types of ensembles are comparable, the uncertainty scores exhibited by ER were found to be dependent on the specific architecture of its base member ('i') and not consistently better than EA. Thus, the challenges posed for the analysis of electron microscopy datasets appear to be better addressed by an ensemble design like EA, as compared to an ensemble design like ER.
Plot2Spectra: an Automatic Spectra Extraction Tool
Jul 06, 2021



Abstract:Different types of spectroscopies, such as X-ray absorption near edge structure (XANES) and Raman spectroscopy, play a very important role in analyzing the characteristics of different materials. In scientific literature, XANES/Raman data are usually plotted in line graphs which is a visually appropriate way to represent the information when the end-user is a human reader. However, such graphs are not conducive to direct programmatic analysis due to the lack of automatic tools. In this paper, we develop a plot digitizer, named Plot2Spectra, to extract data points from spectroscopy graph images in an automatic fashion, which makes it possible for large scale data acquisition and analysis. Specifically, the plot digitizer is a two-stage framework. In the first axis alignment stage, we adopt an anchor-free detector to detect the plot region and then refine the detected bounding boxes with an edge-based constraint to locate the position of two axes. We also apply scene text detector to extract and interpret all tick information below the x-axis. In the second plot data extraction stage, we first employ semantic segmentation to separate pixels belonging to plot lines from the background, and from there, incorporate optical flow constraints to the plot line pixels to assign them to the appropriate line (data instance) they encode. Extensive experiments are conducted to validate the effectiveness of the proposed plot digitizer, which shows that such a tool could help accelerate the discovery and machine learning of materials properties.
EXSCLAIM! -- An automated pipeline for the construction of labeled materials imaging datasets from literature
Mar 19, 2021



Abstract:Due to recent improvements in image resolution and acquisition speed, materials microscopy is experiencing an explosion of published imaging data. The standard publication format, while sufficient for traditional data ingestion scenarios where a select number of images can be critically examined and curated manually, is not conducive to large-scale data aggregation or analysis, hindering data sharing and reuse. Most images in publications are presented as components of a larger figure with their explicit context buried in the main body or caption text, so even if aggregated, collections of images with weak or no digitized contextual labels have limited value. To solve the problem of curating labeled microscopy data from literature, this work introduces the EXSCLAIM! Python toolkit for the automatic EXtraction, Separation, and Caption-based natural Language Annotation of IMages from scientific literature. We highlight the methodology behind the construction of EXSCLAIM! and demonstrate its ability to extract and label open-source scientific images at high volume.
A Two-stage Framework for Compound Figure Separation
Jan 25, 2021



Abstract:Scientific literature contains large volumes of complex, unstructured figures that are compound in nature (i.e. composed of multiple images, graphs, and drawings). Separation of these compound figures is critical for information retrieval from these figures. In this paper, we propose a new strategy for compound figure separation, which decomposes the compound figures into constituent subfigures while preserving the association between the subfigures and their respective caption components. We propose a two-stage framework to address the proposed compound figure separation problem. In particular, the subfigure label detection module detects all subfigure labels in the first stage. Then, in the subfigure detection module, the detected subfigure labels help to detect the subfigures by optimizing the feature selection process and providing the global layout information as extra features. Extensive experiments are conducted to validate the effectiveness and superiority of the proposed framework, which improves the detection precision by 9%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge