Jianguo Wen
Ensemble of Pre-Trained Neural Networks for Segmentation and Quality Detection of Transmission Electron Microscopy Images
Sep 05, 2022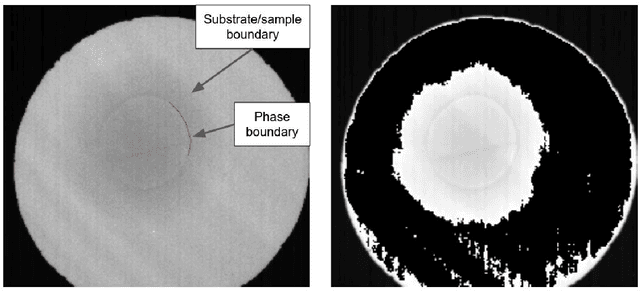

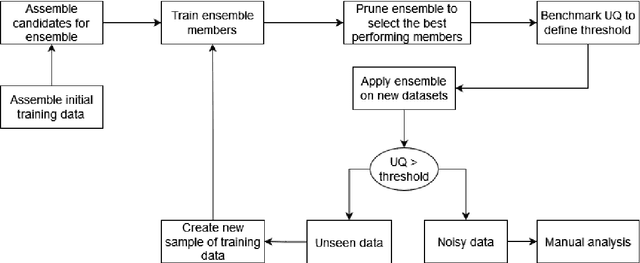
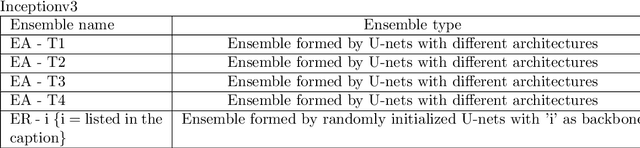
Abstract:Automated analysis of electron microscopy datasets poses multiple challenges, such as limitation in the size of the training dataset, variation in data distribution induced by variation in sample quality and experiment conditions, etc. It is crucial for the trained model to continue to provide acceptable segmentation/classification performance on new data, and quantify the uncertainty associated with its predictions. Among the broad applications of machine learning, various approaches have been adopted to quantify uncertainty, such as Bayesian modeling, Monte Carlo dropout, ensembles, etc. With the aim of addressing the challenges specific to the data domain of electron microscopy, two different types of ensembles of pre-trained neural networks were implemented in this work. The ensembles performed semantic segmentation of ice crystal within a two-phase mixture, thereby tracking its phase transformation to water. The first ensemble (EA) is composed of U-net style networks having different underlying architectures, whereas the second series of ensembles (ER-i) are composed of randomly initialized U-net style networks, wherein each base learner has the same underlying architecture 'i'. The encoders of the base learners were pre-trained on the Imagenet dataset. The performance of EA and ER were evaluated on three different metrics: accuracy, calibration, and uncertainty. It is seen that EA exhibits a greater classification accuracy and is better calibrated, as compared to ER. While the uncertainty quantification of these two types of ensembles are comparable, the uncertainty scores exhibited by ER were found to be dependent on the specific architecture of its base member ('i') and not consistently better than EA. Thus, the challenges posed for the analysis of electron microscopy datasets appear to be better addressed by an ensemble design like EA, as compared to an ensemble design like ER.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge