Arun Baskaran
Ensemble of Pre-Trained Neural Networks for Segmentation and Quality Detection of Transmission Electron Microscopy Images
Sep 05, 2022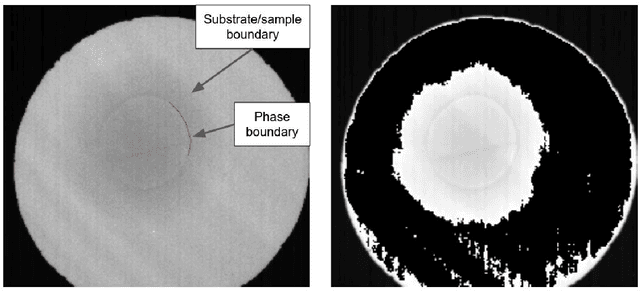

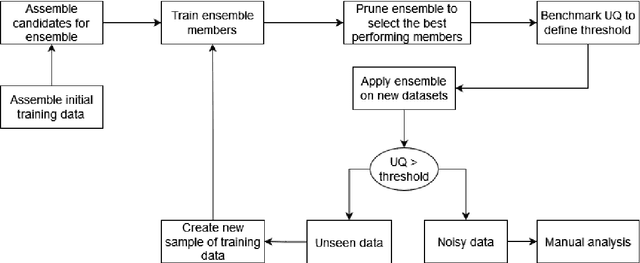
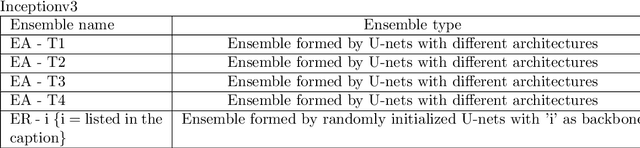
Abstract:Automated analysis of electron microscopy datasets poses multiple challenges, such as limitation in the size of the training dataset, variation in data distribution induced by variation in sample quality and experiment conditions, etc. It is crucial for the trained model to continue to provide acceptable segmentation/classification performance on new data, and quantify the uncertainty associated with its predictions. Among the broad applications of machine learning, various approaches have been adopted to quantify uncertainty, such as Bayesian modeling, Monte Carlo dropout, ensembles, etc. With the aim of addressing the challenges specific to the data domain of electron microscopy, two different types of ensembles of pre-trained neural networks were implemented in this work. The ensembles performed semantic segmentation of ice crystal within a two-phase mixture, thereby tracking its phase transformation to water. The first ensemble (EA) is composed of U-net style networks having different underlying architectures, whereas the second series of ensembles (ER-i) are composed of randomly initialized U-net style networks, wherein each base learner has the same underlying architecture 'i'. The encoders of the base learners were pre-trained on the Imagenet dataset. The performance of EA and ER were evaluated on three different metrics: accuracy, calibration, and uncertainty. It is seen that EA exhibits a greater classification accuracy and is better calibrated, as compared to ER. While the uncertainty quantification of these two types of ensembles are comparable, the uncertainty scores exhibited by ER were found to be dependent on the specific architecture of its base member ('i') and not consistently better than EA. Thus, the challenges posed for the analysis of electron microscopy datasets appear to be better addressed by an ensemble design like EA, as compared to an ensemble design like ER.
The Adoption of Image-Driven Machine Learning for Microstructure Characterization and Materials Design: A Perspective
May 20, 2021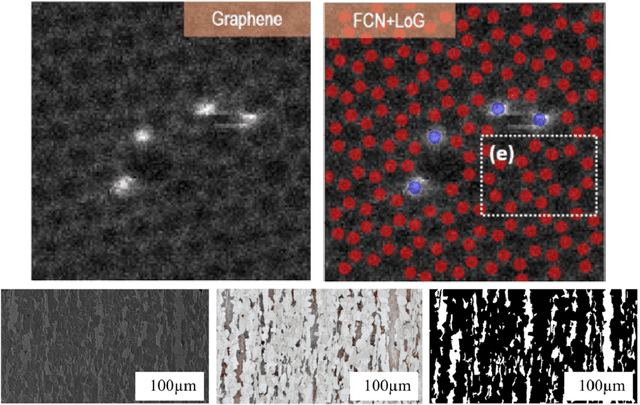
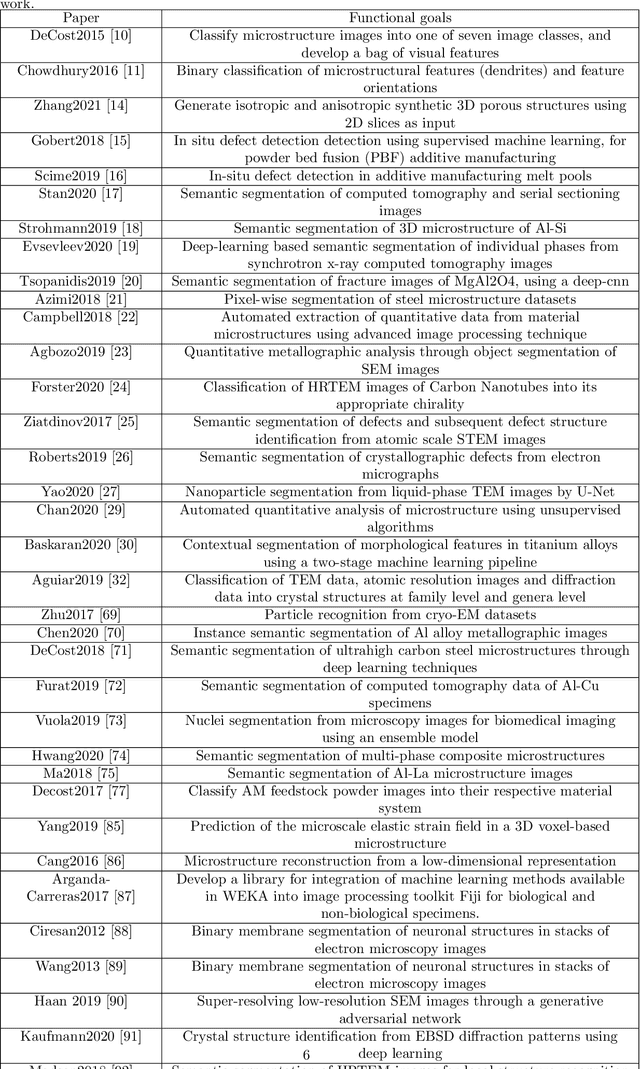
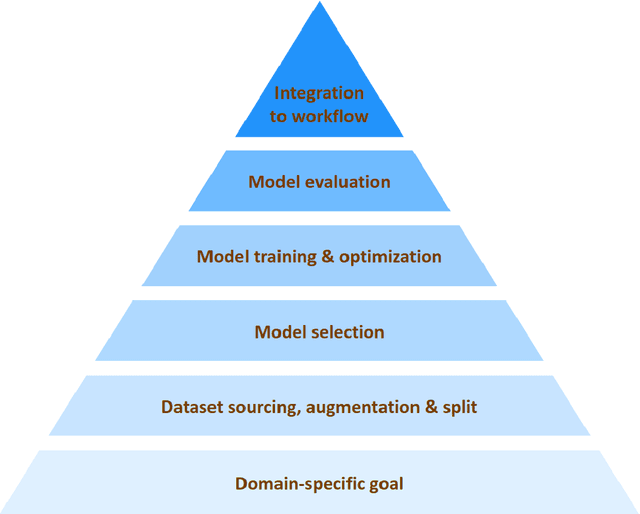
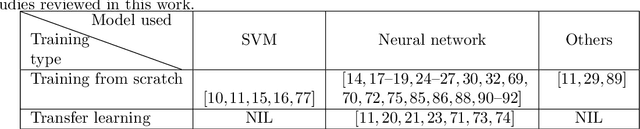
Abstract:The recent surge in the adoption of machine learning techniques for materials design, discovery, and characterization has resulted in an increased interest and application of Image Driven Machine Learning (IDML) approaches. In this work, we review the application of IDML to the field of materials characterization. A hierarchy of six action steps is defined which compartmentalizes a problem statement into well-defined modules. The studies reviewed in this work are analyzed through the decisions adopted by them at each of these steps. Such a review permits a granular assessment of the field, for example the impact of IDML on materials characterization at the nanoscale, the number of images in a typical dataset required to train a semantic segmentation model on electron microscopy images, the prevalence of transfer learning in the domain, etc. Finally, we discuss the importance of interpretability and explainability, and provide an overview of two emerging techniques in the field: semantic segmentation and generative adversarial networks.
Image-driven discriminative and generative machine learning algorithms for establishing microstructure-processing relationships
Jul 27, 2020



Abstract:We investigate methods of microstructure representation for the purpose of predicting processing condition from microstructure image data. A binary alloy (uranium-molybdenum) that is currently under development as a nuclear fuel was studied for the purpose of developing an improved machine learning approach to image recognition, characterization, and building predictive capabilities linking microstructure to processing conditions. Here, we test different microstructure representations and evaluate model performance based on the F1 score. A F1 score of 95.1% was achieved for distinguishing between micrographs corresponding to ten different thermo-mechanical material processing conditions. We find that our newly developed microstructure representation describes image data well, and the traditional approach of utilizing area fractions of different phases is insufficient for distinguishing between multiple classes using a relatively small, imbalanced original data set of 272 images. To explore the applicability of generative methods for supplementing such limited data sets, generative adversarial networks were trained to generate artificial microstructure images. Two different generative networks were trained and tested to assess performance. Challenges and best practices associated with applying machine learning to limited microstructure image data sets is also discussed. Our work has implications for quantitative microstructure analysis, and development of microstructure-processing relationships in limited data sets typical of metallurgical process design studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge