Dane Morgan
Leveraging Vision Capabilities of Multimodal LLMs for Automated Data Extraction from Plots
Mar 16, 2025Abstract:Automated data extraction from research texts has been steadily improving, with the emergence of large language models (LLMs) accelerating progress even further. Extracting data from plots in research papers, however, has been such a complex task that it has predominantly been confined to manual data extraction. We show that current multimodal large language models, with proper instructions and engineered workflows, are capable of accurately extracting data from plots. This capability is inherent to the pretrained models and can be achieved with a chain-of-thought sequence of zero-shot engineered prompts we call PlotExtract, without the need to fine-tune. We demonstrate PlotExtract here and assess its performance on synthetic and published plots. We consider only plots with two axes in this analysis. For plots identified as extractable, PlotExtract finds points with over 90% precision (and around 90% recall) and errors in x and y position of around 5% or lower. These results prove that multimodal LLMs are a viable path for high-throughput data extraction for plots and in many circumstances can replace the current manual methods of data extraction.
A practical guide to machine learning interatomic potentials -- Status and future
Mar 12, 2025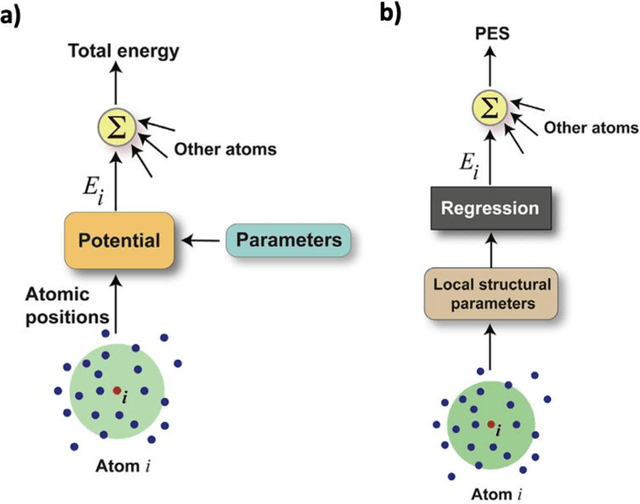

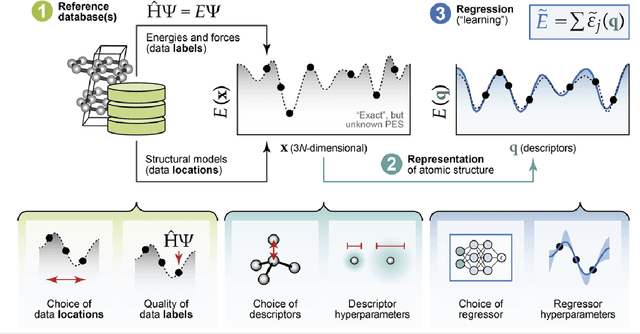
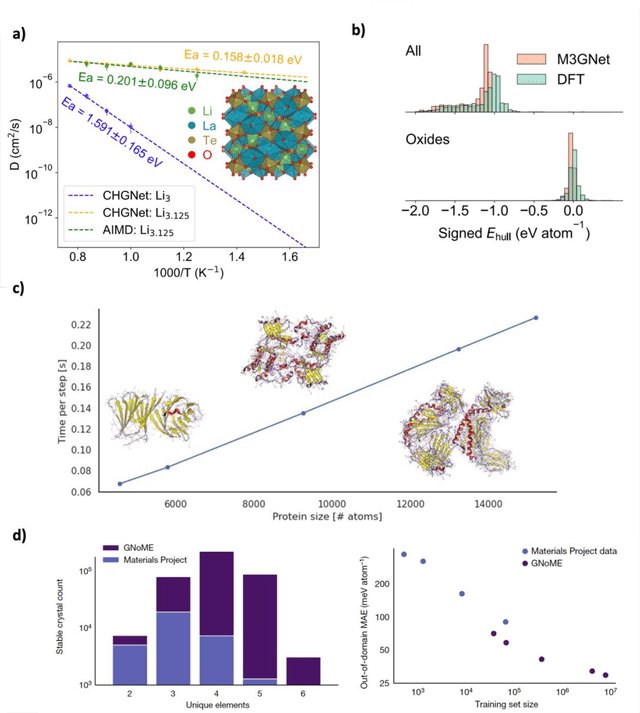
Abstract:The rapid development and large body of literature on machine learning interatomic potentials (MLIPs) can make it difficult to know how to proceed for researchers who are not experts but wish to use these tools. The spirit of this review is to help such researchers by serving as a practical, accessible guide to the state-of-the-art in MLIPs. This review paper covers a broad range of topics related to MLIPs, including (i) central aspects of how and why MLIPs are enablers of many exciting advancements in molecular modeling, (ii) the main underpinnings of different types of MLIPs, including their basic structure and formalism, (iii) the potentially transformative impact of universal MLIPs for both organic and inorganic systems, including an overview of the most recent advances, capabilities, downsides, and potential applications of this nascent class of MLIPs, (iv) a practical guide for estimating and understanding the execution speed of MLIPs, including guidance for users based on hardware availability, type of MLIP used, and prospective simulation size and time, (v) a manual for what MLIP a user should choose for a given application by considering hardware resources, speed requirements, energy and force accuracy requirements, as well as guidance for choosing pre-trained potentials or fitting a new potential from scratch, (vi) discussion around MLIP infrastructure, including sources of training data, pre-trained potentials, and hardware resources for training, (vii) summary of some key limitations of present MLIPs and current approaches to mitigate such limitations, including methods of including long-range interactions, handling magnetic systems, and treatment of excited states, and finally (viii) we finish with some more speculative thoughts on what the future holds for the development and application of MLIPs over the next 3-10+ years.
Predicting Performance of Object Detection Models in Electron Microscopy Using Random Forests
Jan 14, 2025



Abstract:Quantifying prediction uncertainty when applying object detection models to new, unlabeled datasets is critical in applied machine learning. This study introduces an approach to estimate the performance of deep learning-based object detection models for quantifying defects in transmission electron microscopy (TEM) images, focusing on detecting irradiation-induced cavities in TEM images of metal alloys. We developed a random forest regression model that predicts the object detection F1 score, a statistical metric used to evaluate the ability to accurately locate and classify objects of interest. The random forest model uses features extracted from the predictions of the object detection model whose uncertainty is being quantified, enabling fast prediction on new, unlabeled images. The mean absolute error (MAE) for predicting F1 of the trained model on test data is 0.09, and the $R^2$ score is 0.77, indicating there is a significant correlation between the random forest regression model predicted and true defect detection F1 scores. The approach is shown to be robust across three distinct TEM image datasets with varying imaging and material domains. Our approach enables users to estimate the reliability of a defect detection and segmentation model predictions and assess the applicability of the model to their specific datasets, providing valuable information about possible domain shifts and whether the model needs to be fine-tuned or trained on additional data to be maximally effective for the desired use case.
Beyond designer's knowledge: Generating materials design hypotheses via large language models
Sep 10, 2024


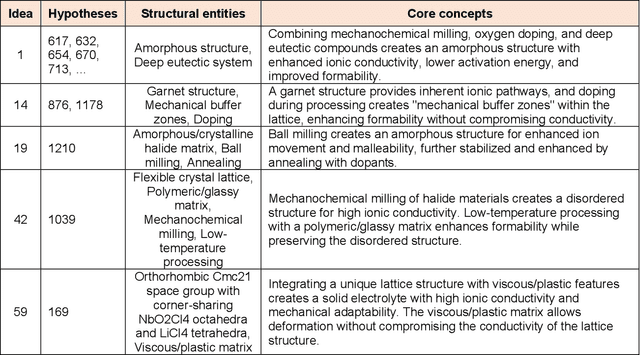
Abstract:Materials design often relies on human-generated hypotheses, a process inherently limited by cognitive constraints such as knowledge gaps and limited ability to integrate and extract knowledge implications, particularly when multidisciplinary expertise is required. This work demonstrates that large language models (LLMs), coupled with prompt engineering, can effectively generate non-trivial materials hypotheses by integrating scientific principles from diverse sources without explicit design guidance by human experts. These include design ideas for high-entropy alloys with superior cryogenic properties and halide solid electrolytes with enhanced ionic conductivity and formability. These design ideas have been experimentally validated in high-impact publications in 2023 not available in the LLM training data, demonstrating the LLM's ability to generate highly valuable and realizable innovative ideas not established in the literature. Our approach primarily leverages materials system charts encoding processing-structure-property relationships, enabling more effective data integration by condensing key information from numerous papers, and evaluation and categorization of numerous hypotheses for human cognition, both through the LLM. This LLM-driven approach opens the door to new avenues of artificial intelligence-driven materials discovery by accelerating design, democratizing innovation, and expanding capabilities beyond the designer's direct knowledge.
Regression with Large Language Models for Materials and Molecular Property Prediction
Sep 09, 2024Abstract:We demonstrate the ability of large language models (LLMs) to perform material and molecular property regression tasks, a significant deviation from the conventional LLM use case. We benchmark the Large Language Model Meta AI (LLaMA) 3 on several molecular properties in the QM9 dataset and 24 materials properties. Only composition-based input strings are used as the model input and we fine tune on only the generative loss. We broadly find that LLaMA 3, when fine-tuned using the SMILES representation of molecules, provides useful regression results which can rival standard materials property prediction models like random forest or fully connected neural networks on the QM9 dataset. Not surprisingly, LLaMA 3 errors are 5-10x higher than those of the state-of-the-art models that were trained using far more granular representation of molecules (e.g., atom types and their coordinates) for the same task. Interestingly, LLaMA 3 provides improved predictions compared to GPT-3.5 and GPT-4o. This work highlights the versatility of LLMs, suggesting that LLM-like generative models can potentially transcend their traditional applications to tackle complex physical phenomena, thus paving the way for future research and applications in chemistry, materials science and other scientific domains.
Accelerating Domain-Aware Electron Microscopy Analysis Using Deep Learning Models with Synthetic Data and Image-Wide Confidence Scoring
Aug 02, 2024Abstract:The integration of machine learning (ML) models enhances the efficiency, affordability, and reliability of feature detection in microscopy, yet their development and applicability are hindered by the dependency on scarce and often flawed manually labeled datasets and a lack of domain awareness. We addressed these challenges by creating a physics-based synthetic image and data generator, resulting in a machine learning model that achieves comparable precision (0.86), recall (0.63), F1 scores (0.71), and engineering property predictions (R2=0.82) to a model trained on human-labeled data. We enhanced both models by using feature prediction confidence scores to derive an image-wide confidence metric, enabling simple thresholding to eliminate ambiguous and out-of-domain images resulting in performance boosts of 5-30% with a filtering-out rate of 25%. Our study demonstrates that synthetic data can eliminate human reliance in ML and provides a means for domain awareness in cases where many feature detections per image are needed.
Determining Domain of Machine Learning Models using Kernel Density Estimates: Applications in Materials Property Prediction
May 28, 2024



Abstract:Knowledge of the domain of applicability of a machine learning model is essential to ensuring accurate and reliable model predictions. In this work, we develop a new approach of assessing model domain and demonstrate that our approach provides accurate and meaningful designation of in-domain versus out-of-domain when applied across multiple model types and material property data sets. Our approach assesses the distance between a test and training data point in feature space by using kernel density estimation and shows that this distance provides an effective tool for domain determination. We show that chemical groups considered unrelated based on established chemical knowledge exhibit significant dissimilarities by our measure. We also show that high measures of dissimilarity are associated with poor model performance (i.e., high residual magnitudes) and poor estimates of model uncertainty (i.e., unreliable uncertainty estimation). Automated tools are provided to enable researchers to establish acceptable dissimilarity thresholds to identify whether new predictions of their own machine learning models are in-domain versus out-of-domain.
Accelerating Ensemble Error Bar Prediction with Single Models Fits
Apr 15, 2024Abstract:Ensemble models can be used to estimate prediction uncertainties in machine learning models. However, an ensemble of N models is approximately N times more computationally demanding compared to a single model when it is used for inference. In this work, we explore fitting a single model to predicted ensemble error bar data, which allows us to estimate uncertainties without the need for a full ensemble. Our approach is based on three models: Model A for predictive accuracy, Model $A_{E}$ for traditional ensemble-based error bar prediction, and Model B, fit to data from Model $A_{E}$, to be used for predicting the values of $A_{E}$ but with only one model evaluation. Model B leverages synthetic data augmentation to estimate error bars efficiently. This approach offers a highly flexible method of uncertainty quantification that can approximate that of ensemble methods but only requires a single extra model evaluation over Model A during inference. We assess this approach on a set of problems in materials science.
Extracting Accurate Materials Data from Research Papers with Conversational Language Models and Prompt Engineering -- Example of ChatGPT
Mar 07, 2023Abstract:There has been a growing effort to replace hand extraction of data from research papers with automated data extraction based on natural language processing (NLP), language models (LMs), and recently, large language models (LLMs). Although these methods enable efficient extraction of data from large sets of research papers, they require a significant amount of up-front effort, expertise, and coding. In this work we propose the ChatExtract method that can fully automate very accurate data extraction with essentially no initial effort or background using an advanced conversational LLM (or AI). ChatExtract consists of a set of engineered prompts applied to a conversational LLM that both identify sentences with data, extract data, and assure its correctness through a series of follow-up questions. These follow-up questions address a critical challenge associated with LLMs - their tendency to provide factually inaccurate responses. ChatExtract can be applied with any conversational LLMs and yields very high quality data extraction. In tests on materials data we find precision and recall both over 90% from the best conversational LLMs, likely rivaling or exceeding human accuracy in many cases. We demonstrate that the exceptional performance is enabled by the information retention in a conversational model combined with purposeful redundancy and introducing uncertainty through follow-up prompts. These results suggest that approaches similar to ChatExtract, due to their simplicity, transferability and accuracy are likely to replace other methods of data extraction in the near future.
Flexible, Model-Agnostic Method for Materials Data Extraction from Text Using General Purpose Language Models
Feb 09, 2023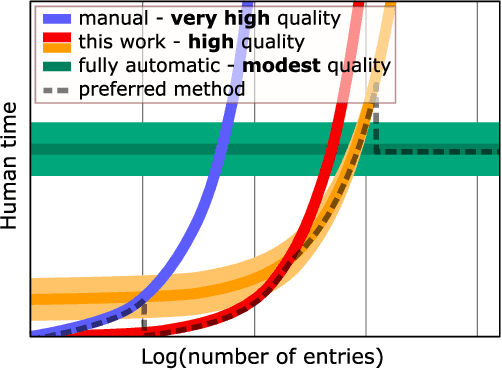
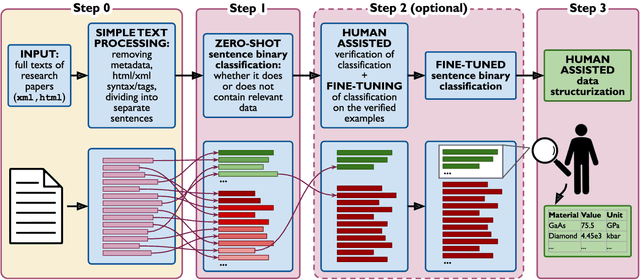
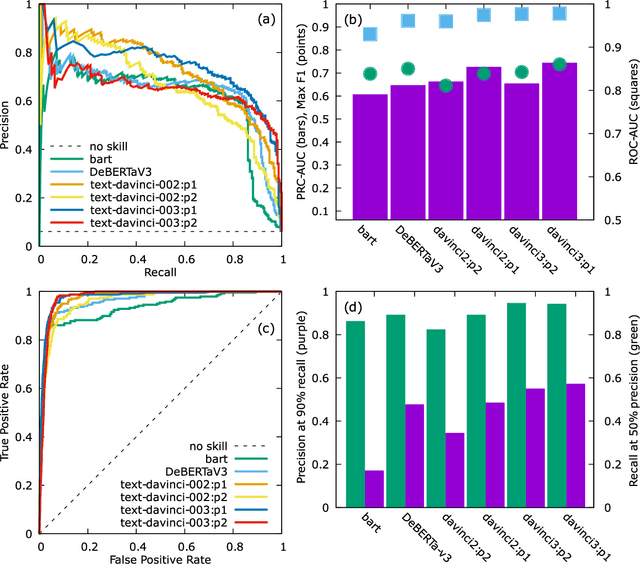
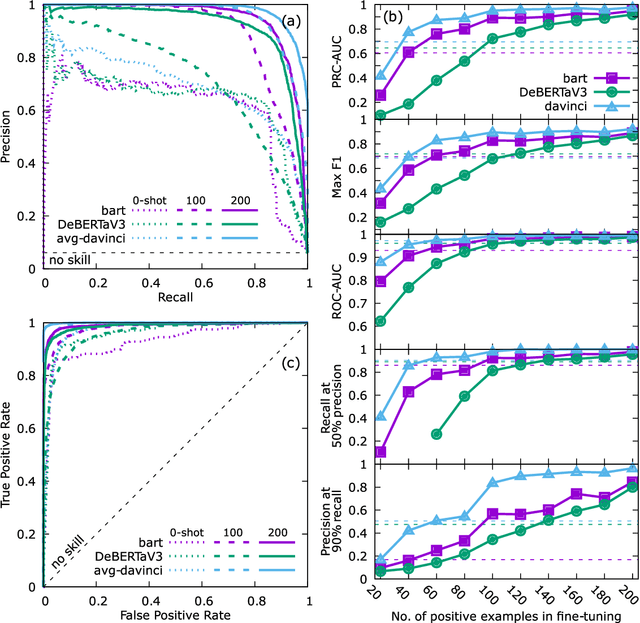
Abstract:Accurate and comprehensive material databases extracted from research papers are critical for materials science and engineering but require significant human effort to develop. In this paper we present a simple method of extracting materials data from full texts of research papers suitable for quickly developing modest-sized databases. The method requires minimal to no coding, prior knowledge about the extracted property, or model training, and provides high recall and almost perfect precision in the resultant database. The method is fully automated except for one human-assisted step, which typically requires just a few hours of human labor. The method builds on top of natural language processing and large general language models but can work with almost any such model. The language models GPT-3/3.5, bart and DeBERTaV3 are evaluated here for comparison. We provide a detailed detailed analysis of the methods performance in extracting bulk modulus data, obtaining up to 90% precision at 96% recall, depending on the amount of human effort involved. We then demonstrate the methods broader effectiveness by developing a database of critical cooling rates for metallic glasses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge