Nicholas Krämer
Numerically robust Gaussian state estimation with singular observation noise
Mar 13, 2025Abstract:This article proposes numerically robust algorithms for Gaussian state estimation with singular observation noise. Our approach combines a series of basis changes with Bayes' rule, transforming the singular estimation problem into a nonsingular one with reduced state dimension. In addition to ensuring low runtime and numerical stability, our proposal facilitates marginal-likelihood computations and Gauss-Markov representations of the posterior process. We analyse the proposed method's computational savings and numerical robustness and validate our findings in a series of simulations.
Adaptive Probabilistic ODE Solvers Without Adaptive Memory Requirements
Oct 14, 2024Abstract:Despite substantial progress in recent years, probabilistic solvers with adaptive step sizes can still not solve memory-demanding differential equations -- unless we care only about a single point in time (which is far too restrictive; we want the whole time series). Counterintuitively, the culprit is the adaptivity itself: Its unpredictable memory demands easily exceed our machine's capabilities, making our simulations fail unexpectedly and without warning. Still, dropping adaptivity would abandon years of progress, which can't be the answer. In this work, we solve this conundrum. We develop an adaptive probabilistic solver with fixed memory demands building on recent developments in robust state estimation. Switching to our method (i) eliminates memory issues for long time series, (ii) accelerates simulations by orders of magnitude through unlocking just-in-time compilation, and (iii) makes adaptive probabilistic solvers compatible with scientific computing in JAX.
Numerically Robust Fixed-Point Smoothing Without State Augmentation
Sep 30, 2024Abstract:Practical implementations of Gaussian smoothing algorithms have received a great deal of attention in the last 60 years. However, almost all work focuses on estimating complete time series (''fixed-interval smoothing'', $\mathcal{O}(K)$ memory) through variations of the Rauch--Tung--Striebel smoother, rarely on estimating the initial states (''fixed-point smoothing'', $\mathcal{O}(1)$ memory). Since fixed-point smoothing is a crucial component of algorithms for dynamical systems with unknown initial conditions, we close this gap by introducing a new formulation of a Gaussian fixed-point smoother. In contrast to prior approaches, our perspective admits a numerically robust Cholesky-based form (without downdates) and avoids state augmentation, which would needlessly inflate the state-space model and reduce the numerical practicality of any fixed-point smoother code. The experiments demonstrate how a JAX implementation of our algorithm matches the runtime of the fastest methods and the robustness of the most robust techniques while existing implementations must always sacrifice one for the other.
A tutorial on automatic differentiation with complex numbers
Sep 10, 2024Abstract:Automatic differentiation is everywhere, but there exists only minimal documentation of how it works in complex arithmetic beyond stating "derivatives in $\mathbb{C}^d$" $\cong$ "derivatives in $\mathbb{R}^{2d}$" and, at best, shallow references to Wirtinger calculus. Unfortunately, the equivalence $\mathbb{C}^d \cong \mathbb{R}^{2d}$ becomes insufficient as soon as we need to derive custom gradient rules, e.g., to avoid differentiating "through" expensive linear algebra functions or differential equation simulators. To combat such a lack of documentation, this article surveys forward- and reverse-mode automatic differentiation with complex numbers, covering topics such as Wirtinger derivatives, a modified chain rule, and different gradient conventions while explicitly avoiding holomorphicity and the Cauchy--Riemann equations (which would be far too restrictive). To be precise, we will derive, explain, and implement a complex version of Jacobian-vector and vector-Jacobian products almost entirely with linear algebra without relying on complex analysis or differential geometry. This tutorial is a call to action, for users and developers alike, to take complex values seriously when implementing custom gradient propagation rules -- the manuscript explains how.
Gradients of Functions of Large Matrices
May 27, 2024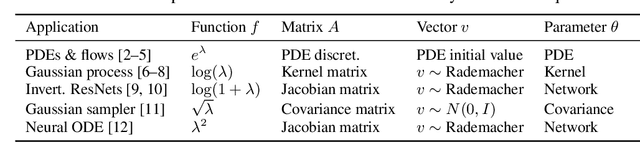



Abstract:Tuning scientific and probabilistic machine learning models -- for example, partial differential equations, Gaussian processes, or Bayesian neural networks -- often relies on evaluating functions of matrices whose size grows with the data set or the number of parameters. While the state-of-the-art for evaluating these quantities is almost always based on Lanczos and Arnoldi iterations, the present work is the first to explain how to differentiate these workhorses of numerical linear algebra efficiently. To get there, we derive previously unknown adjoint systems for Lanczos and Arnoldi iterations, implement them in JAX, and show that the resulting code can compete with Diffrax when it comes to differentiating PDEs, GPyTorch for selecting Gaussian process models and beats standard factorisation methods for calibrating Bayesian neural networks. All this is achieved without any problem-specific code optimisation. Find the code at https://github.com/pnkraemer/experiments-lanczos-adjoints and install the library with pip install matfree.
Approximate Bayesian Neural Operators: Uncertainty Quantification for Parametric PDEs
Aug 02, 2022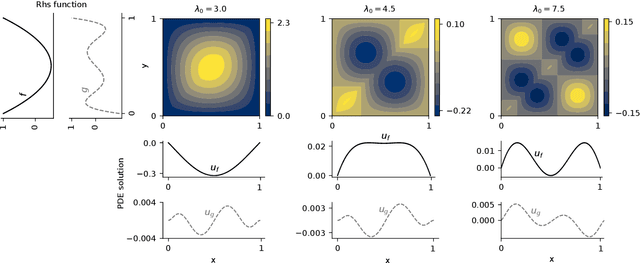
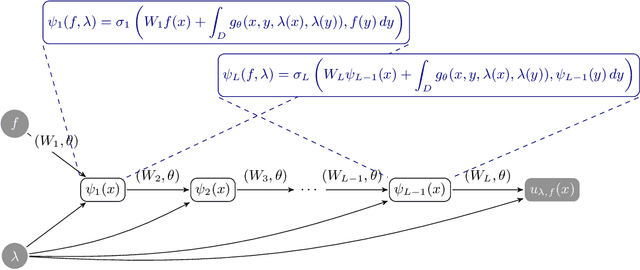
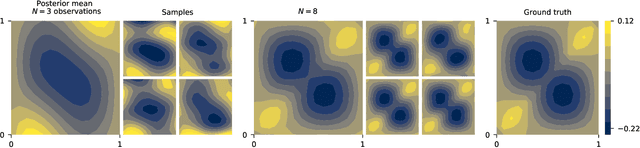
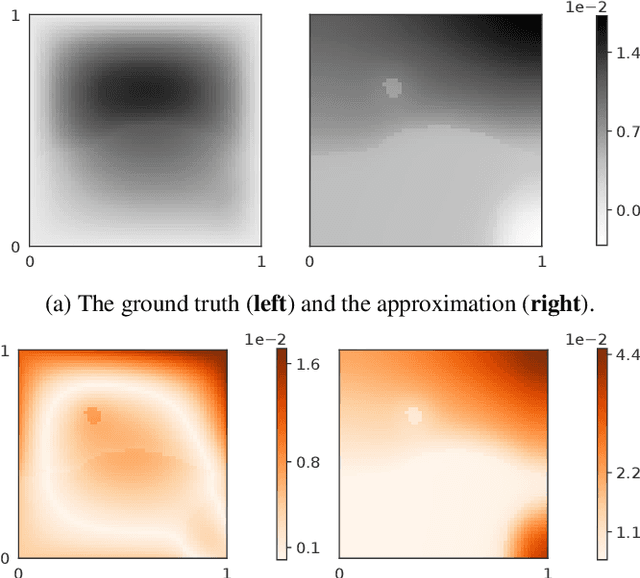
Abstract:Neural operators are a type of deep architecture that learns to solve (i.e. learns the nonlinear solution operator of) partial differential equations (PDEs). The current state of the art for these models does not provide explicit uncertainty quantification. This is arguably even more of a problem for this kind of tasks than elsewhere in machine learning, because the dynamical systems typically described by PDEs often exhibit subtle, multiscale structure that makes errors hard to spot by humans. In this work, we first provide a mathematically detailed Bayesian formulation of the ''shallow'' (linear) version of neural operators in the formalism of Gaussian processes. We then extend this analytic treatment to general deep neural operators using approximate methods from Bayesian deep learning. We extend previous results on neural operators by providing them with uncertainty quantification. As a result, our approach is able to identify cases, and provide structured uncertainty estimates, where the neural operator fails to predict well.
ProbNum: Probabilistic Numerics in Python
Dec 03, 2021
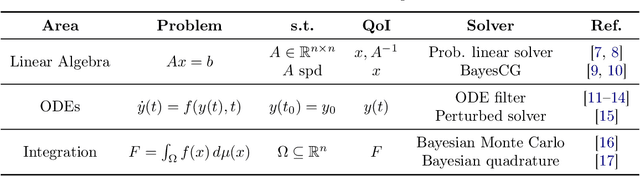
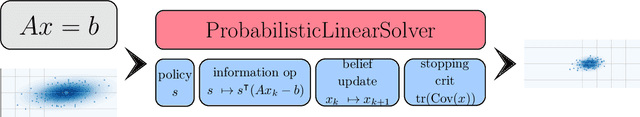
Abstract:Probabilistic numerical methods (PNMs) solve numerical problems via probabilistic inference. They have been developed for linear algebra, optimization, integration and differential equation simulation. PNMs naturally incorporate prior information about a problem and quantify uncertainty due to finite computational resources as well as stochastic input. In this paper, we present ProbNum: a Python library providing state-of-the-art probabilistic numerical solvers. ProbNum enables custom composition of PNMs for specific problem classes via a modular design as well as wrappers for off-the-shelf use. Tutorials, documentation, developer guides and benchmarks are available online at www.probnum.org.
Probabilistic Numerical Method of Lines for Time-Dependent Partial Differential Equations
Oct 22, 2021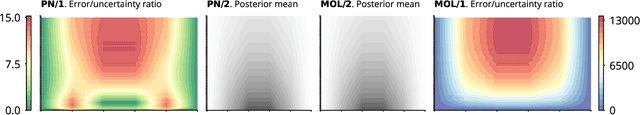
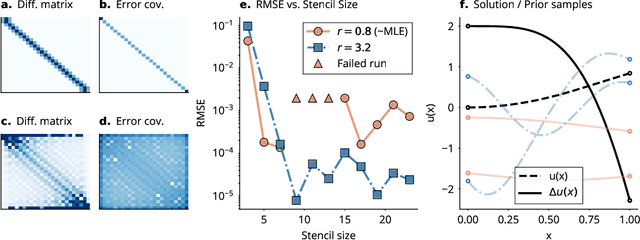
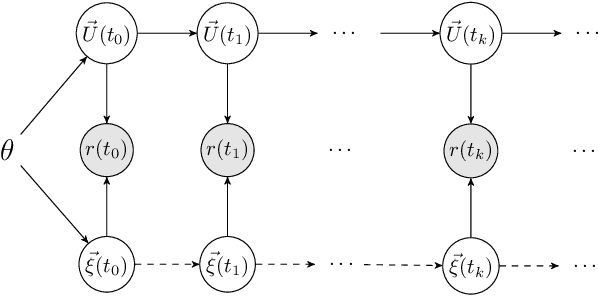
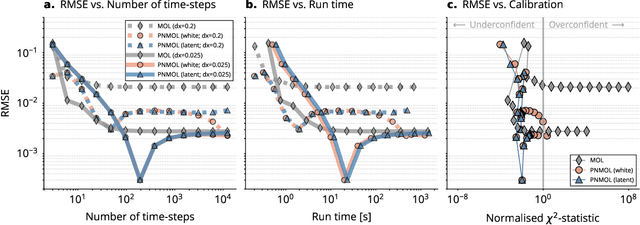
Abstract:This work develops a class of probabilistic algorithms for the numerical solution of nonlinear, time-dependent partial differential equations (PDEs). Current state-of-the-art PDE solvers treat the space- and time-dimensions separately, serially, and with black-box algorithms, which obscures the interactions between spatial and temporal approximation errors and misguides the quantification of the overall error. To fix this issue, we introduce a probabilistic version of a technique called method of lines. The proposed algorithm begins with a Gaussian process interpretation of finite difference methods, which then interacts naturally with filtering-based probabilistic ordinary differential equation (ODE) solvers because they share a common language: Bayesian inference. Joint quantification of space- and time-uncertainty becomes possible without losing the performance benefits of well-tuned ODE solvers. Thereby, we extend the toolbox of probabilistic programs for differential equation simulation to PDEs.
Probabilistic ODE Solutions in Millions of Dimensions
Oct 22, 2021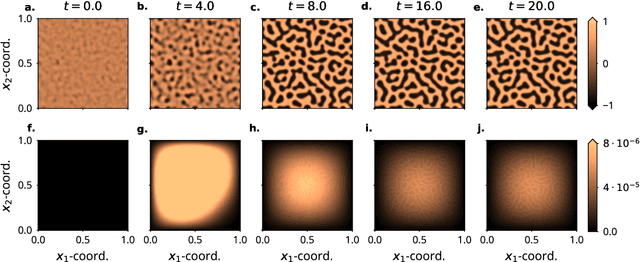
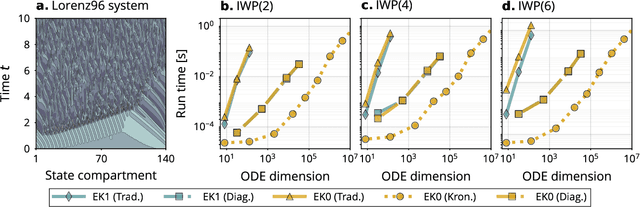
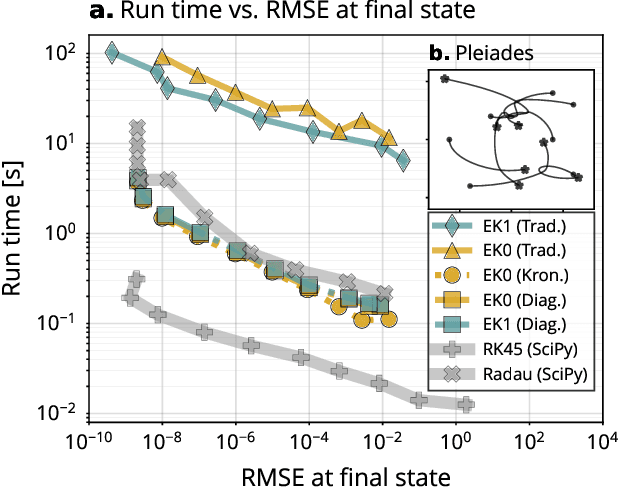
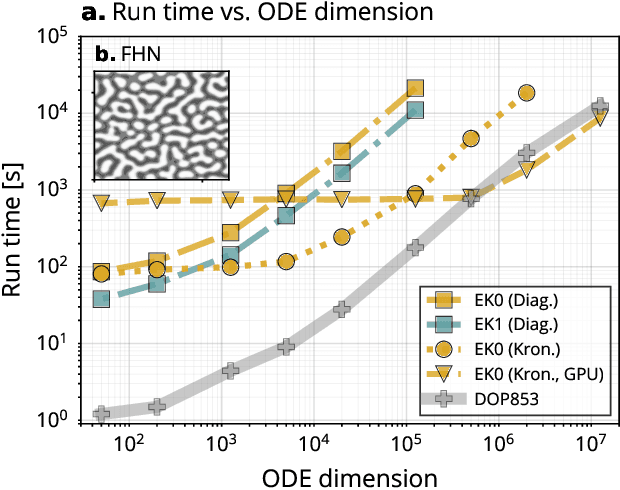
Abstract:Probabilistic solvers for ordinary differential equations (ODEs) have emerged as an efficient framework for uncertainty quantification and inference on dynamical systems. In this work, we explain the mathematical assumptions and detailed implementation schemes behind solving {high-dimensional} ODEs with a probabilistic numerical algorithm. This has not been possible before due to matrix-matrix operations in each solver step, but is crucial for scientifically relevant problems -- most importantly, the solution of discretised {partial} differential equations. In a nutshell, efficient high-dimensional probabilistic ODE solutions build either on independence assumptions or on Kronecker structure in the prior model. We evaluate the resulting efficiency on a range of problems, including the probabilistic numerical simulation of a differential equation with millions of dimensions.
Linear-Time Probabilistic Solutions of Boundary Value Problems
Jun 14, 2021

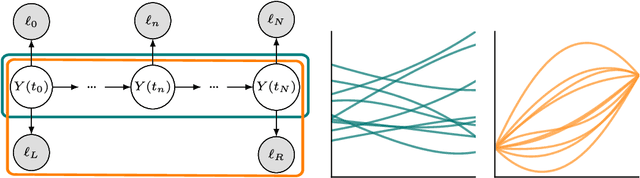

Abstract:We propose a fast algorithm for the probabilistic solution of boundary value problems (BVPs), which are ordinary differential equations subject to boundary conditions. In contrast to previous work, we introduce a Gauss--Markov prior and tailor it specifically to BVPs, which allows computing a posterior distribution over the solution in linear time, at a quality and cost comparable to that of well-established, non-probabilistic methods. Our model further delivers uncertainty quantification, mesh refinement, and hyperparameter adaptation. We demonstrate how these practical considerations positively impact the efficiency of the scheme. Altogether, this results in a practically usable probabilistic BVP solver that is (in contrast to non-probabilistic algorithms) natively compatible with other parts of the statistical modelling tool-chain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge