Gradients of Functions of Large Matrices
Paper and Code
May 27, 2024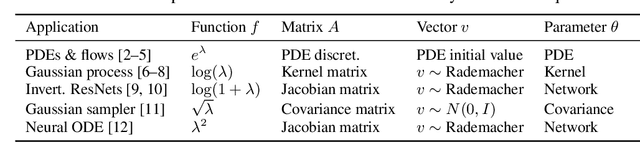



Tuning scientific and probabilistic machine learning models -- for example, partial differential equations, Gaussian processes, or Bayesian neural networks -- often relies on evaluating functions of matrices whose size grows with the data set or the number of parameters. While the state-of-the-art for evaluating these quantities is almost always based on Lanczos and Arnoldi iterations, the present work is the first to explain how to differentiate these workhorses of numerical linear algebra efficiently. To get there, we derive previously unknown adjoint systems for Lanczos and Arnoldi iterations, implement them in JAX, and show that the resulting code can compete with Diffrax when it comes to differentiating PDEs, GPyTorch for selecting Gaussian process models and beats standard factorisation methods for calibrating Bayesian neural networks. All this is achieved without any problem-specific code optimisation. Find the code at https://github.com/pnkraemer/experiments-lanczos-adjoints and install the library with pip install matfree.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge