Mulan Jin
Breast Cancer Immunohistochemical Image Generation: a Benchmark Dataset and Challenge Review
May 05, 2023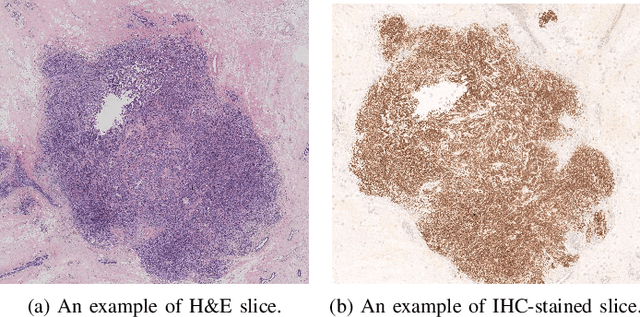
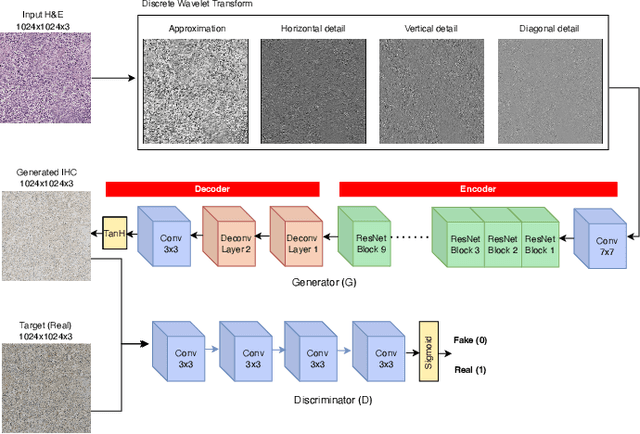
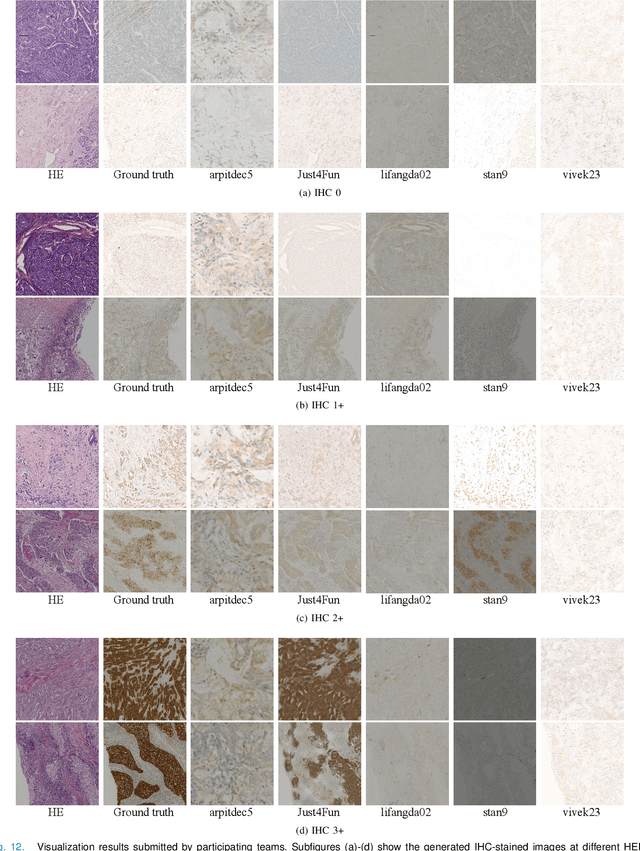
Abstract:For invasive breast cancer, immunohistochemical (IHC) techniques are often used to detect the expression level of human epidermal growth factor receptor-2 (HER2) in breast tissue to formulate a precise treatment plan. From the perspective of saving manpower, material and time costs, directly generating IHC-stained images from hematoxylin and eosin (H&E) stained images is a valuable research direction. Therefore, we held the breast cancer immunohistochemical image generation challenge, aiming to explore novel ideas of deep learning technology in pathological image generation and promote research in this field. The challenge provided registered H&E and IHC-stained image pairs, and participants were required to use these images to train a model that can directly generate IHC-stained images from corresponding H&E-stained images. We selected and reviewed the five highest-ranking methods based on their PSNR and SSIM metrics, while also providing overviews of the corresponding pipelines and implementations. In this paper, we further analyze the current limitations in the field of breast cancer immunohistochemical image generation and forecast the future development of this field. We hope that the released dataset and the challenge will inspire more scholars to jointly study higher-quality IHC-stained image generation.
BCI: Breast Cancer Immunohistochemical Image Generation through Pyramid Pix2pix
Apr 25, 2022



Abstract:The evaluation of human epidermal growth factor receptor 2 (HER2) expression is essential to formulate a precise treatment for breast cancer. The routine evaluation of HER2 is conducted with immunohistochemical techniques (IHC), which is very expensive. Therefore, for the first time, we propose a breast cancer immunohistochemical (BCI) benchmark attempting to synthesize IHC data directly with the paired hematoxylin and eosin (HE) stained images. The dataset contains 4870 registered image pairs, covering a variety of HER2 expression levels. Based on BCI, as a minor contribution, we further build a pyramid pix2pix image generation method, which achieves better HE to IHC translation results than the other current popular algorithms. Extensive experiments demonstrate that BCI poses new challenges to the existing image translation research. Besides, BCI also opens the door for future pathology studies in HER2 expression evaluation based on the synthesized IHC images. BCI dataset can be downloaded from https://bupt-ai-cz.github.io/BCI.
Predicting Axillary Lymph Node Metastasis in Early Breast Cancer Using Deep Learning on Primary Tumor Biopsy Slides
Dec 12, 2021



Abstract:Objectives: To develop and validate a deep learning (DL)-based primary tumor biopsy signature for predicting axillary lymph node (ALN) metastasis preoperatively in early breast cancer (EBC) patients with clinically negative ALN. Methods: A total of 1,058 EBC patients with pathologically confirmed ALN status were enrolled from May 2010 to August 2020. A DL core-needle biopsy (DL-CNB) model was built on the attention-based multiple instance-learning (AMIL) framework to predict ALN status utilizing the DL features, which were extracted from the cancer areas of digitized whole-slide images (WSIs) of breast CNB specimens annotated by two pathologists. Accuracy, sensitivity, specificity, receiver operating characteristic (ROC) curves, and areas under the ROC curve (AUCs) were analyzed to evaluate our model. Results: The best-performing DL-CNB model with VGG16_BN as the feature extractor achieved an AUC of 0.816 (95% confidence interval (CI): 0.758, 0.865) in predicting positive ALN metastasis in the independent test cohort. Furthermore, our model incorporating the clinical data, which was called DL-CNB+C, yielded the best accuracy of 0.831 (95%CI: 0.775, 0.878), especially for patients younger than 50 years (AUC: 0.918, 95%CI: 0.825, 0.971). The interpretation of DL-CNB model showed that the top signatures most predictive of ALN metastasis were characterized by the nucleus features including density ($p$ = 0.015), circumference ($p$ = 0.009), circularity ($p$ = 0.010), and orientation ($p$ = 0.012). Conclusion: Our study provides a novel DL-based biomarker on primary tumor CNB slides to predict the metastatic status of ALN preoperatively for patients with EBC. The codes and dataset are available at https://github.com/bupt-ai-cz/BALNMP
* Accepted by Frontiers in Oncology, for more details, please see https://github.com/bupt-ai-cz/BALNMP
Hard Sample Aware Noise Robust Learning for Histopathology Image Classification
Dec 05, 2021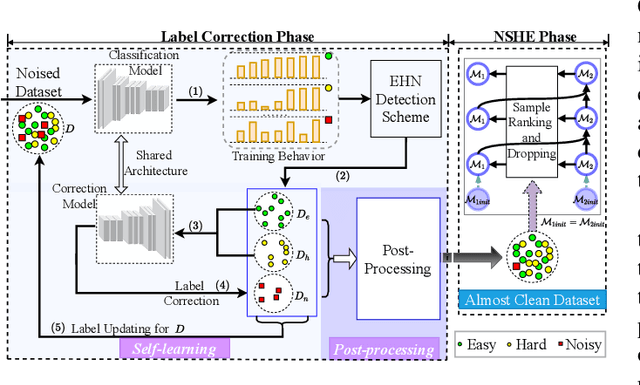
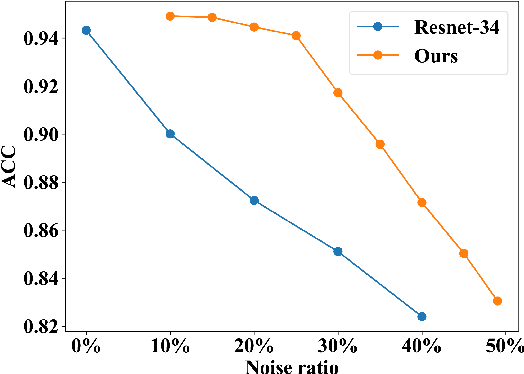
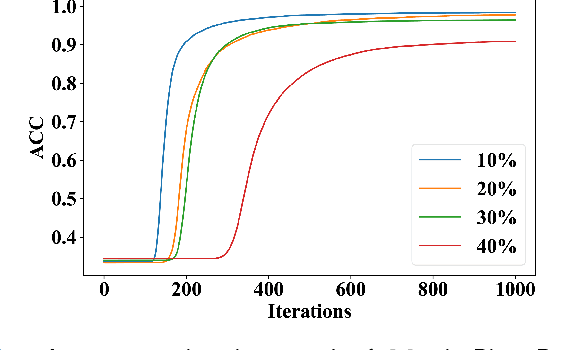
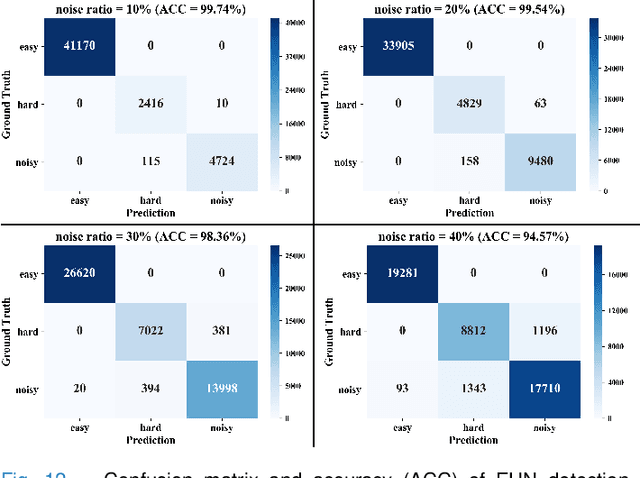
Abstract:Deep learning-based histopathology image classification is a key technique to help physicians in improving the accuracy and promptness of cancer diagnosis. However, the noisy labels are often inevitable in the complex manual annotation process, and thus mislead the training of the classification model. In this work, we introduce a novel hard sample aware noise robust learning method for histopathology image classification. To distinguish the informative hard samples from the harmful noisy ones, we build an easy/hard/noisy (EHN) detection model by using the sample training history. Then we integrate the EHN into a self-training architecture to lower the noise rate through gradually label correction. With the obtained almost clean dataset, we further propose a noise suppressing and hard enhancing (NSHE) scheme to train the noise robust model. Compared with the previous works, our method can save more clean samples and can be directly applied to the real-world noisy dataset scenario without using a clean subset. Experimental results demonstrate that the proposed scheme outperforms the current state-of-the-art methods in both the synthetic and real-world noisy datasets. The source code and data are available at https://github.com/bupt-ai-cz/HSA-NRL/.
Multi-level colonoscopy malignant tissue detection with adversarial CAC-UNet
Jun 30, 2020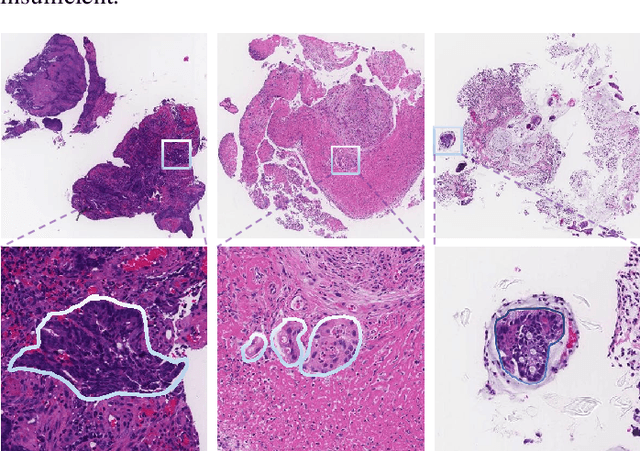

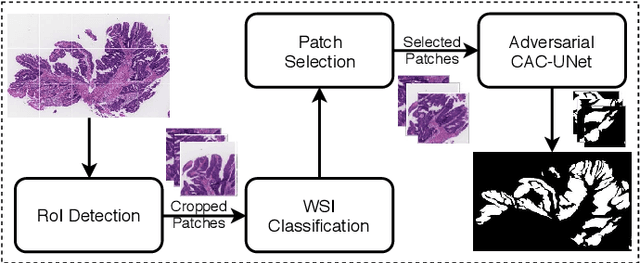
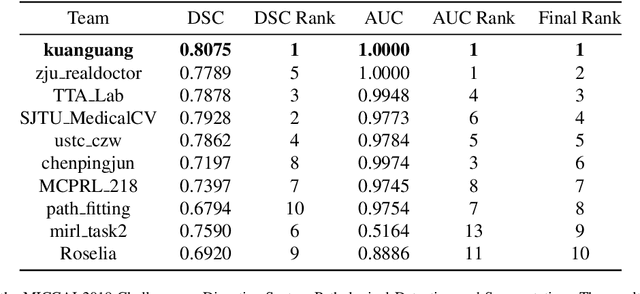
Abstract:The automatic and objective medical diagnostic model can be valuable to achieve early cancer detection, and thus reducing the mortality rate. In this paper, we propose a highly efficient multi-level malignant tissue detection through the designed adversarial CAC-UNet. A patch-level model with a pre-prediction strategy and a malignancy area guided label smoothing is adopted to remove the negative WSIs, with which to lower the risk of false positive detection. For the selected key patches by multi-model ensemble, an adversarial context-aware and appearance consistency UNet (CAC-UNet) is designed to achieve robust segmentation. In CAC-UNet, mirror designed discriminators are able to seamlessly fuse the whole feature maps of the skillfully designed powerful backbone network without any information loss. Besides, a mask prior is further added to guide the accurate segmentation mask prediction through an extra mask-domain discriminator. The proposed scheme achieves the best results in MICCAI DigestPath2019 challenge on colonoscopy tissue segmentation and classification task. The full implementation details and the trained models are available at https://github.com/Raykoooo/CAC-UNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge