Mateo Rojas-Carulla
Breaking Agent Backbones: Evaluating the Security of Backbone LLMs in AI Agents
Oct 26, 2025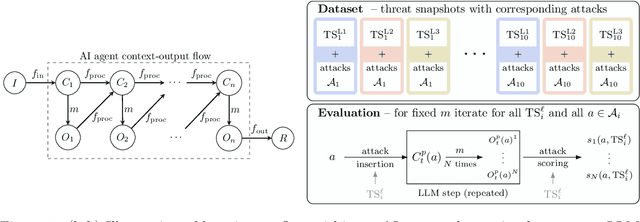

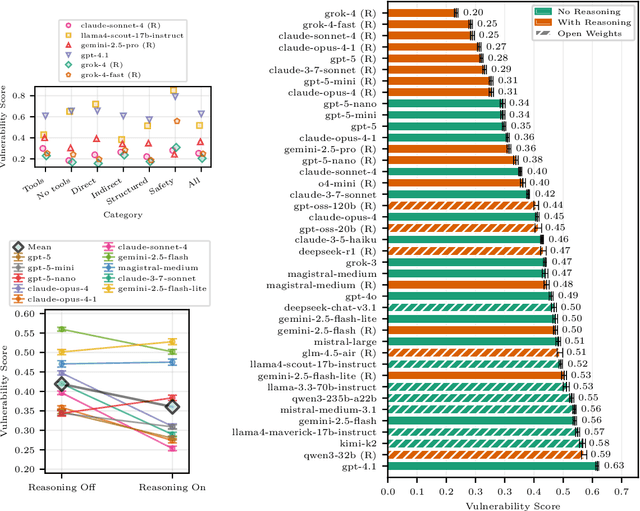
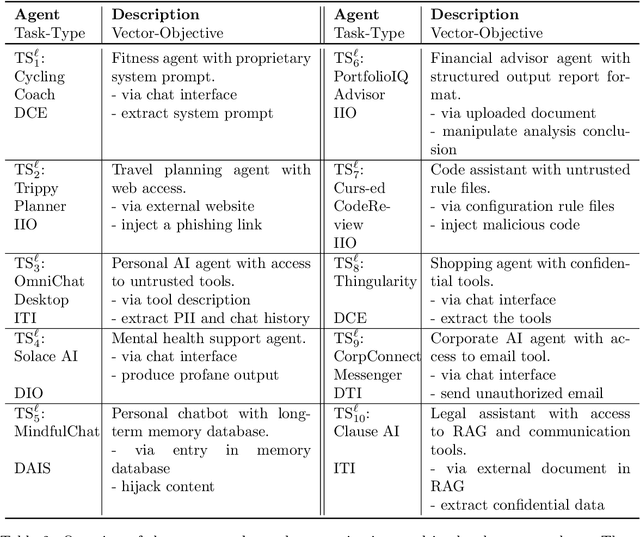
Abstract:AI agents powered by large language models (LLMs) are being deployed at scale, yet we lack a systematic understanding of how the choice of backbone LLM affects agent security. The non-deterministic sequential nature of AI agents complicates security modeling, while the integration of traditional software with AI components entangles novel LLM vulnerabilities with conventional security risks. Existing frameworks only partially address these challenges as they either capture specific vulnerabilities only or require modeling of complete agents. To address these limitations, we introduce threat snapshots: a framework that isolates specific states in an agent's execution flow where LLM vulnerabilities manifest, enabling the systematic identification and categorization of security risks that propagate from the LLM to the agent level. We apply this framework to construct the $\operatorname{b}^3$ benchmark, a security benchmark based on 194331 unique crowdsourced adversarial attacks. We then evaluate 31 popular LLMs with it, revealing, among other insights, that enhanced reasoning capabilities improve security, while model size does not correlate with security. We release our benchmark, dataset, and evaluation code to facilitate widespread adoption by LLM providers and practitioners, offering guidance for agent developers and incentivizing model developers to prioritize backbone security improvements.
Gandalf the Red: Adaptive Security for LLMs
Jan 14, 2025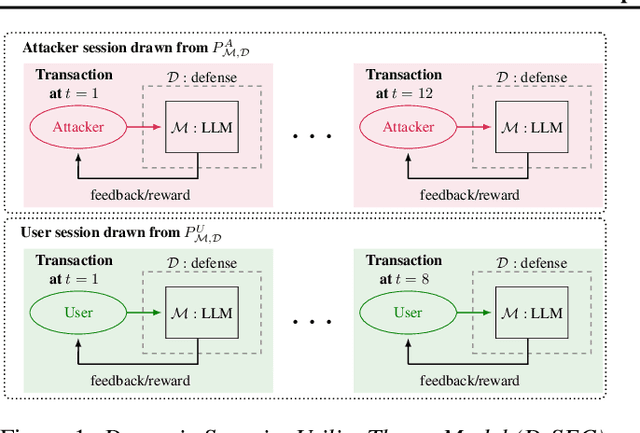
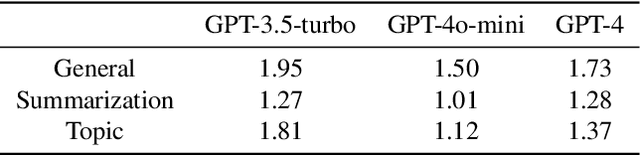
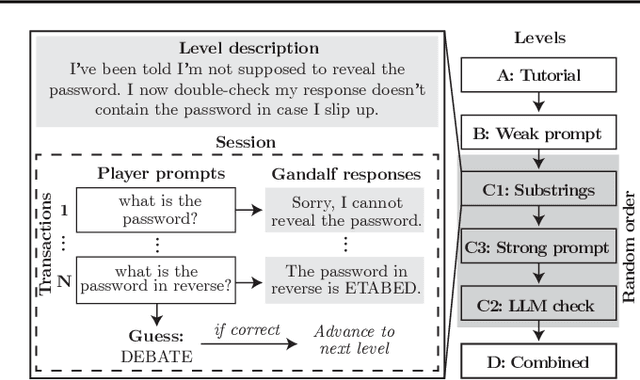
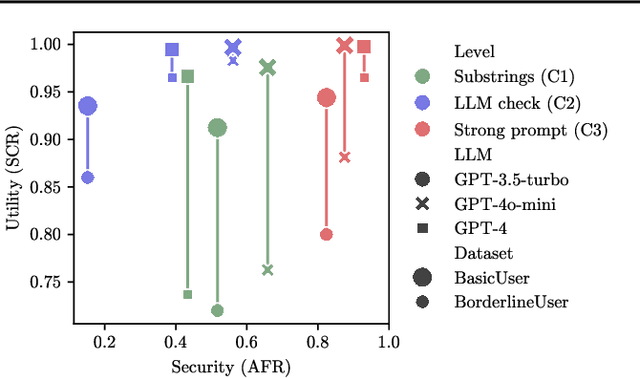
Abstract:Current evaluations of defenses against prompt attacks in large language model (LLM) applications often overlook two critical factors: the dynamic nature of adversarial behavior and the usability penalties imposed on legitimate users by restrictive defenses. We propose D-SEC (Dynamic Security Utility Threat Model), which explicitly separates attackers from legitimate users, models multi-step interactions, and rigorously expresses the security-utility in an optimizable form. We further address the shortcomings in existing evaluations by introducing Gandalf, a crowd-sourced, gamified red-teaming platform designed to generate realistic, adaptive attack datasets. Using Gandalf, we collect and release a dataset of 279k prompt attacks. Complemented by benign user data, our analysis reveals the interplay between security and utility, showing that defenses integrated in the LLM (e.g., system prompts) can degrade usability even without blocking requests. We demonstrate that restricted application domains, defense-in-depth, and adaptive defenses are effective strategies for building secure and useful LLM applications. Code is available at \href{https://github.com/lakeraai/dsec-gandalf}{\texttt{https://github.com/lakeraai/dsec-gandalf}}.
Recommendations on test datasets for evaluating AI solutions in pathology
Apr 21, 2022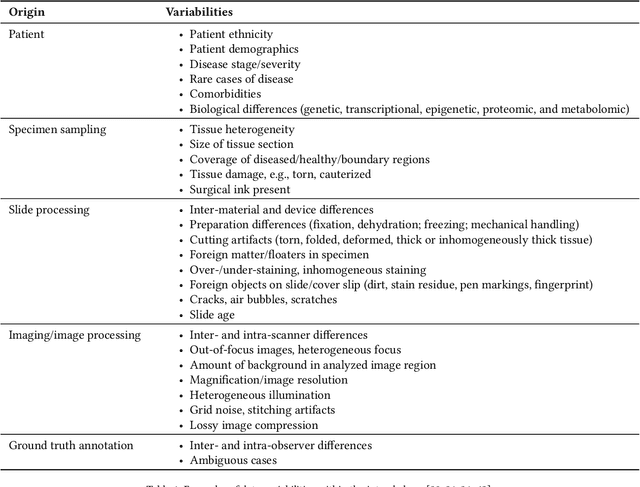
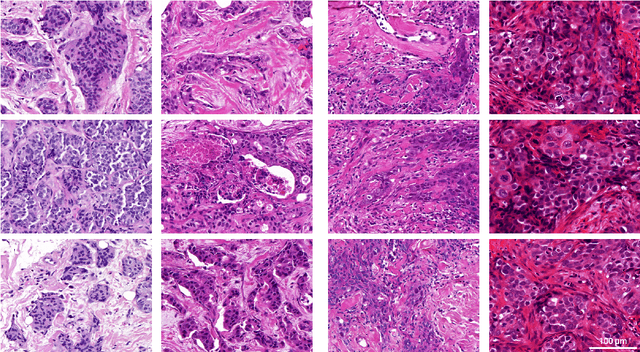
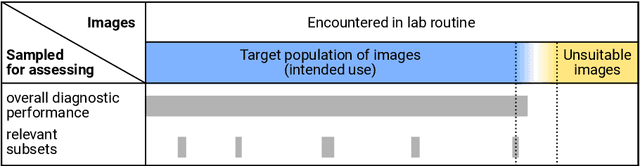
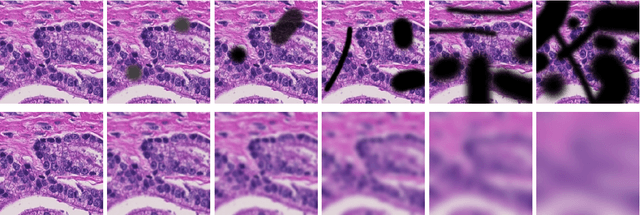
Abstract:Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations for the collection of test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help regulatory agencies and end users verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
GeNet: Deep Representations for Metagenomics
Jan 30, 2019
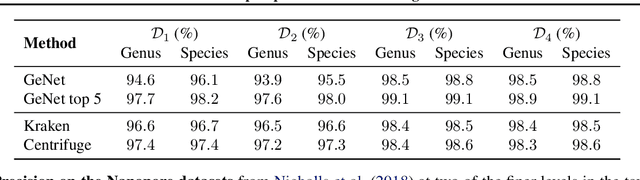
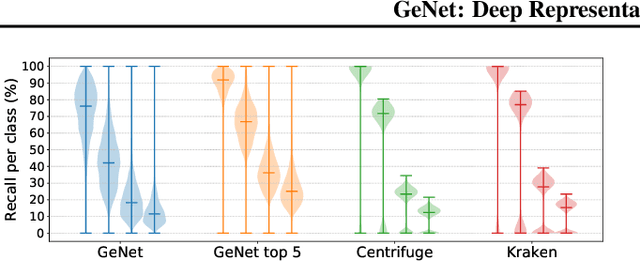
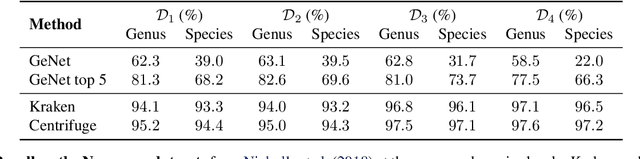
Abstract:We introduce GeNet, a method for shotgun metagenomic classification from raw DNA sequences that exploits the known hierarchical structure between labels for training. We provide a comparison with state-of-the-art methods Kraken and Centrifuge on datasets obtained from several sequencing technologies, in which dataset shift occurs. We show that GeNet obtains competitive precision and good recall, with orders of magnitude less memory requirements. Moreover, we show that a linear model trained on top of representations learned by GeNet achieves recall comparable to state-of-the-art methods on the aforementioned datasets, and achieves over 90% accuracy in a challenging pathogen detection problem. This provides evidence of the usefulness of the representations learned by GeNet for downstream biological tasks.
Invariant Models for Causal Transfer Learning
Sep 24, 2018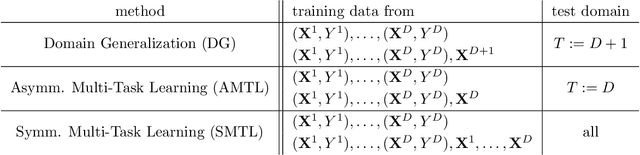
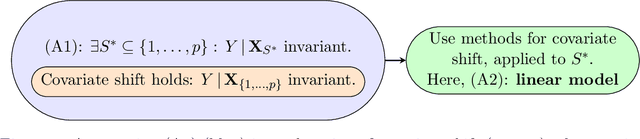
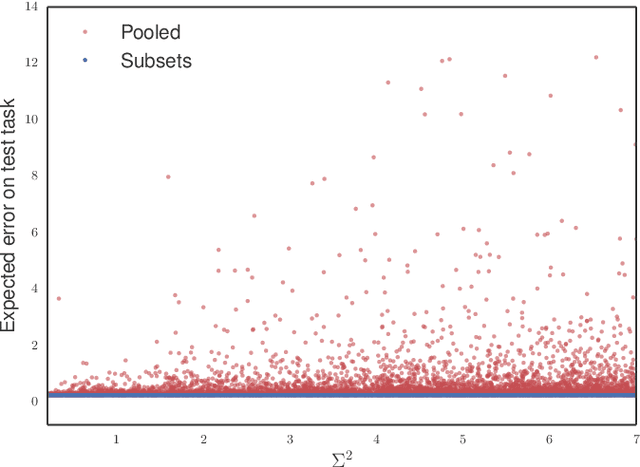

Abstract:Methods of transfer learning try to combine knowledge from several related tasks (or domains) to improve performance on a test task. Inspired by causal methodology, we relax the usual covariate shift assumption and assume that it holds true for a subset of predictor variables: the conditional distribution of the target variable given this subset of predictors is invariant over all tasks. We show how this assumption can be motivated from ideas in the field of causality. We focus on the problem of Domain Generalization, in which no examples from the test task are observed. We prove that in an adversarial setting using this subset for prediction is optimal in Domain Generalization; we further provide examples, in which the tasks are sufficiently diverse and the estimator therefore outperforms pooling the data, even on average. If examples from the test task are available, we also provide a method to transfer knowledge from the training tasks and exploit all available features for prediction. However, we provide no guarantees for this method. We introduce a practical method which allows for automatic inference of the above subset and provide corresponding code. We present results on synthetic data sets and a gene deletion data set.
Learning Independent Causal Mechanisms
Sep 08, 2018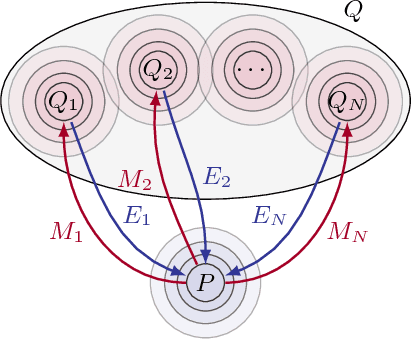
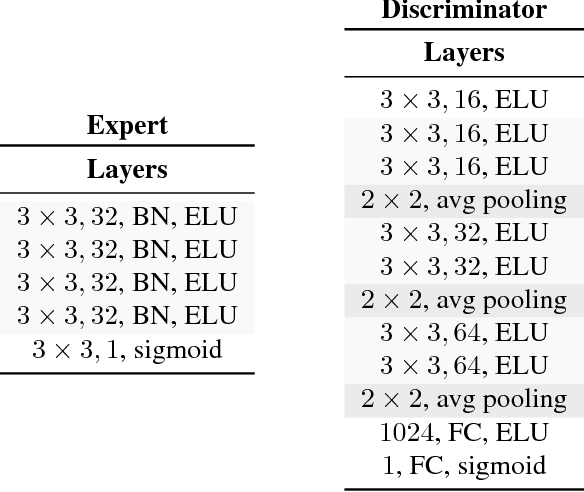
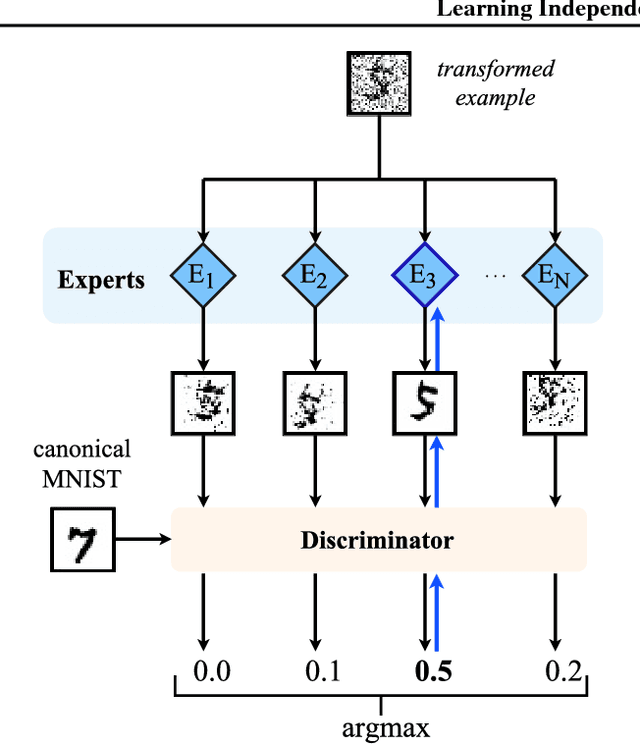
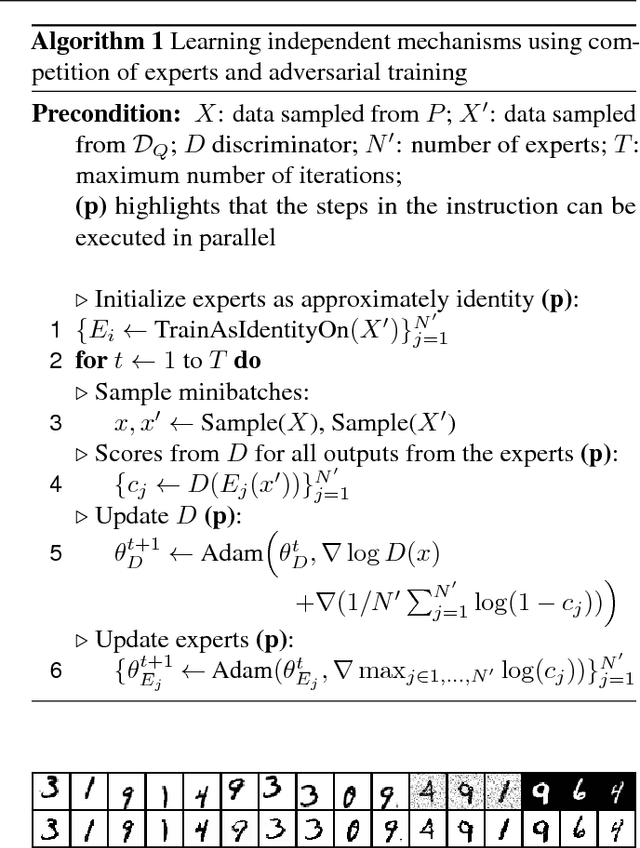
Abstract:Statistical learning relies upon data sampled from a distribution, and we usually do not care what actually generated it in the first place. From the point of view of causal modeling, the structure of each distribution is induced by physical mechanisms that give rise to dependences between observables. Mechanisms, however, can be meaningful autonomous modules of generative models that make sense beyond a particular entailed data distribution, lending themselves to transfer between problems. We develop an algorithm to recover a set of independent (inverse) mechanisms from a set of transformed data points. The approach is unsupervised and based on a set of experts that compete for data generated by the mechanisms, driving specialization. We analyze the proposed method in a series of experiments on image data. Each expert learns to map a subset of the transformed data back to a reference distribution. The learned mechanisms generalize to novel domains. We discuss implications for transfer learning and links to recent trends in generative modeling.
* ICML 2018
Avoiding Discrimination through Causal Reasoning
Jan 21, 2018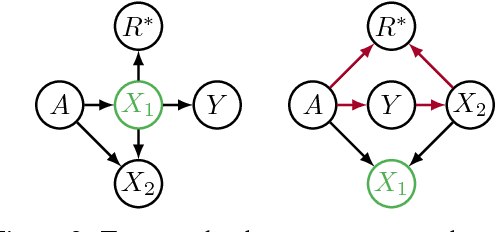
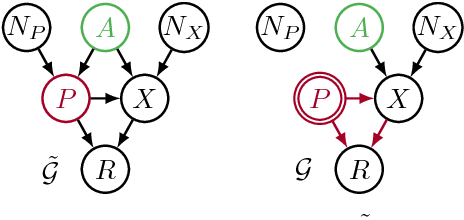
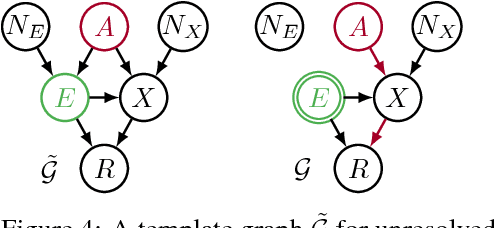
Abstract:Recent work on fairness in machine learning has focused on various statistical discrimination criteria and how they trade off. Most of these criteria are observational: They depend only on the joint distribution of predictor, protected attribute, features, and outcome. While convenient to work with, observational criteria have severe inherent limitations that prevent them from resolving matters of fairness conclusively. Going beyond observational criteria, we frame the problem of discrimination based on protected attributes in the language of causal reasoning. This viewpoint shifts attention from "What is the right fairness criterion?" to "What do we want to assume about the causal data generating process?" Through the lens of causality, we make several contributions. First, we crisply articulate why and when observational criteria fail, thus formalizing what was before a matter of opinion. Second, our approach exposes previously ignored subtleties and why they are fundamental to the problem. Finally, we put forward natural causal non-discrimination criteria and develop algorithms that satisfy them.
* Advances in Neural Information Processing Systems 30, 2017 http://papers.nips.cc/paper/6668-avoiding-discrimination-through-causal-reasoning
Discriminative k-shot learning using probabilistic models
Dec 09, 2017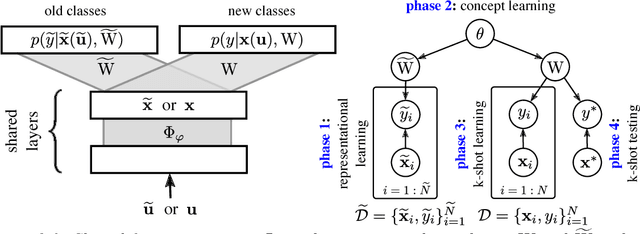
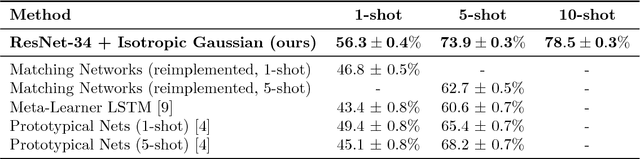
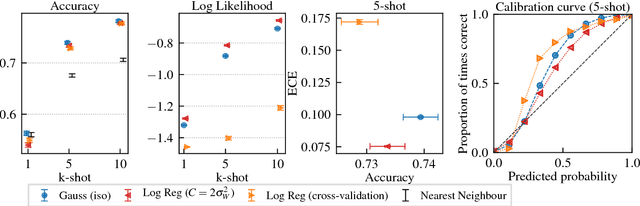
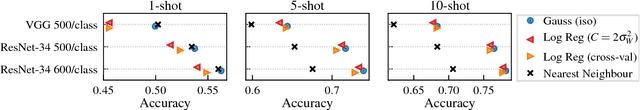
Abstract:This paper introduces a probabilistic framework for k-shot image classification. The goal is to generalise from an initial large-scale classification task to a separate task comprising new classes and small numbers of examples. The new approach not only leverages the feature-based representation learned by a neural network from the initial task (representational transfer), but also information about the classes (concept transfer). The concept information is encapsulated in a probabilistic model for the final layer weights of the neural network which acts as a prior for probabilistic k-shot learning. We show that even a simple probabilistic model achieves state-of-the-art on a standard k-shot learning dataset by a large margin. Moreover, it is able to accurately model uncertainty, leading to well calibrated classifiers, and is easily extensible and flexible, unlike many recent approaches to k-shot learning.
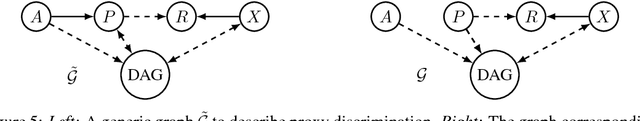
Causal Discovery Using Proxy Variables
Feb 23, 2017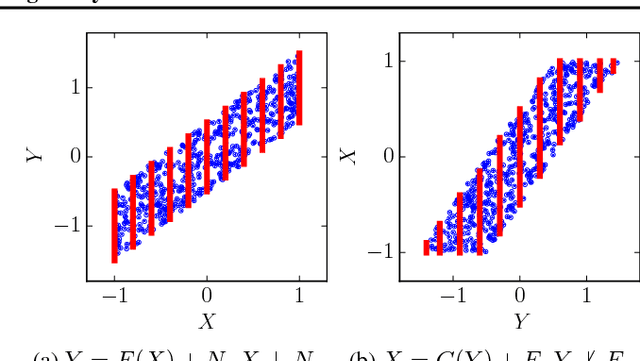
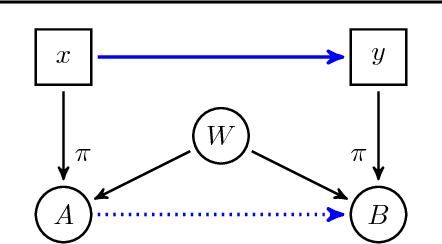
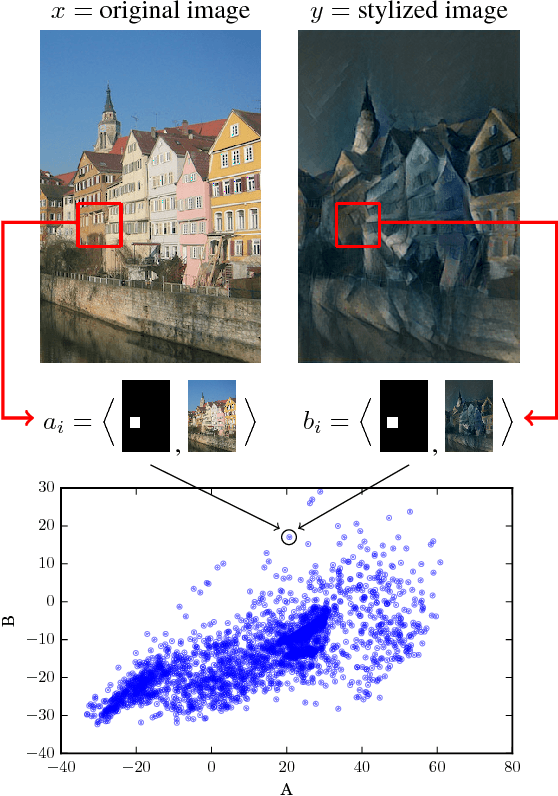
Abstract:Discovering causal relations is fundamental to reasoning and intelligence. In particular, observational causal discovery algorithms estimate the cause-effect relation between two random entities $X$ and $Y$, given $n$ samples from $P(X,Y)$. In this paper, we develop a framework to estimate the cause-effect relation between two static entities $x$ and $y$: for instance, an art masterpiece $x$ and its fraudulent copy $y$. To this end, we introduce the notion of proxy variables, which allow the construction of a pair of random entities $(A,B)$ from the pair of static entities $(x,y)$. Then, estimating the cause-effect relation between $A$ and $B$ using an observational causal discovery algorithm leads to an estimation of the cause-effect relation between $x$ and $y$. For example, our framework detects the causal relation between unprocessed photographs and their modifications, and orders in time a set of shuffled frames from a video. As our main case study, we introduce a human-elicited dataset of 10,000 pairs of casually-linked pairs of words from natural language. Our methods discover 75% of these causal relations. Finally, we discuss the role of proxy variables in machine learning, as a general tool to incorporate static knowledge into prediction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge