Martin Schwartz
SERA-H: Beyond Native Sentinel Spatial Limits for High-Resolution Canopy Height Mapping
Dec 19, 2025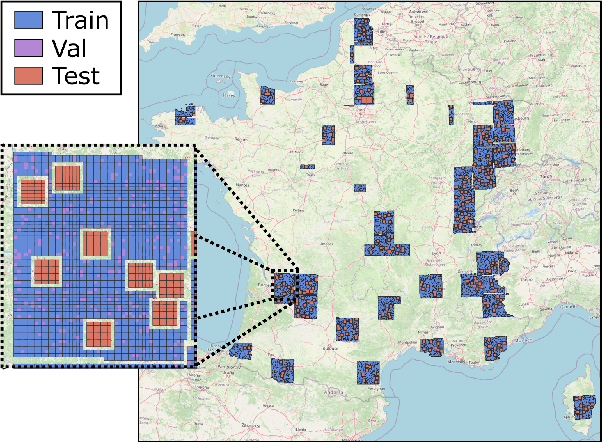
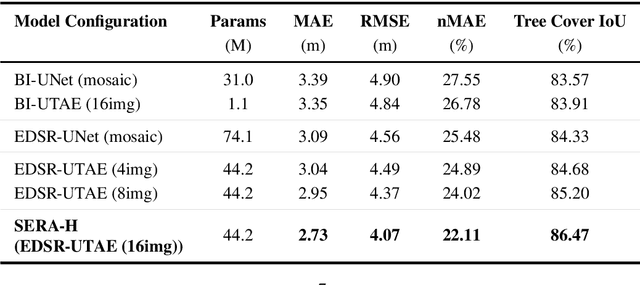
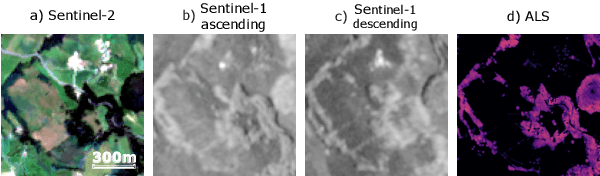
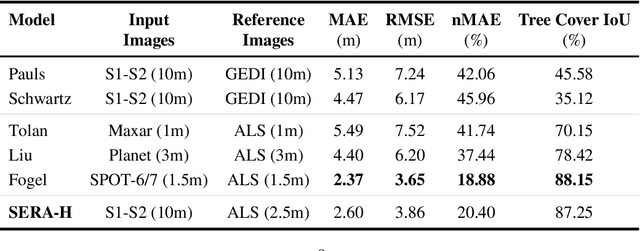
Abstract:High-resolution mapping of canopy height is essential for forest management and biodiversity monitoring. Although recent studies have led to the advent of deep learning methods using satellite imagery to predict height maps, these approaches often face a trade-off between data accessibility and spatial resolution. To overcome these limitations, we present SERA-H, an end-to-end model combining a super-resolution module (EDSR) and temporal attention encoding (UTAE). Trained under the supervision of high-density LiDAR data (ALS), our model generates 2.5 m resolution height maps from freely available Sentinel-1 and Sentinel-2 (10 m) time series data. Evaluated on an open-source benchmark dataset in France, SERA-H, with a MAE of 2.6 m and a coefficient of determination of 0.82, not only outperforms standard Sentinel-1/2 baselines but also achieves performance comparable to or better than methods relying on commercial very high-resolution imagery (SPOT-6/7, PlanetScope, Maxar). These results demonstrate that combining high-resolution supervision with the spatiotemporal information embedded in time series enables the reconstruction of details beyond the input sensors' native resolution. SERA-H opens the possibility of freely mapping forests with high revisit frequency, achieving accuracy comparable to that of costly commercial imagery. The source code is available at https://github.com/ThomasBoudras/SERA-H#
FORMSpoT: A Decade of Tree-Level, Country-Scale Forest Monitoring
Dec 18, 2025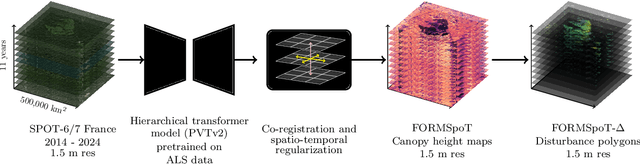
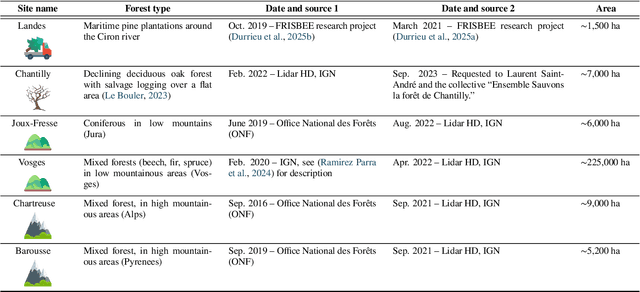
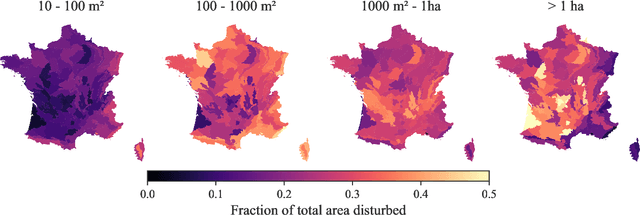
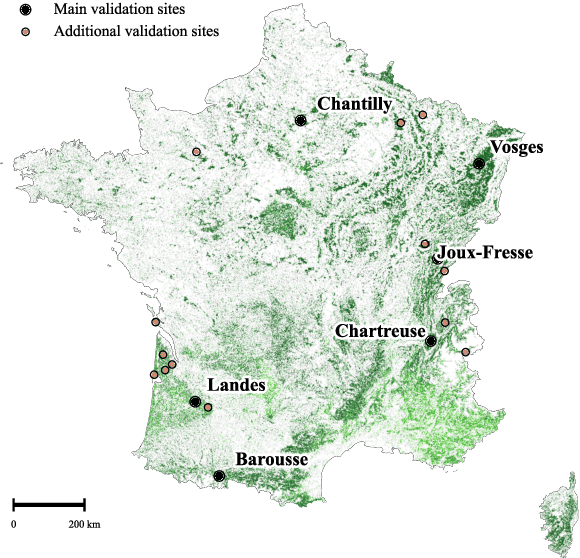
Abstract:The recent decline of the European forest carbon sink highlights the need for spatially explicit and frequently updated forest monitoring tools. Yet, existing satellite-based disturbance products remain too coarse to detect changes at the scale of individual trees, typically below 100 m$^{2}$. Here, we introduce FORMSpoT (Forest Mapping with SPOT Time series), a decade-long (2014-2024) nationwide mapping of forest canopy height at 1.5 m resolution, together with annual disturbance polygons (FORMSpoT-$Δ$) covering mainland France. Canopy heights were derived from annual SPOT-6/7 composites using a hierarchical transformer model (PVTv2) trained on high-resolution airborne laser scanning (ALS) data. To enable robust change detection across heterogeneous acquisitions, we developed a dedicated post-processing pipeline combining co-registration and spatio-temporal total variation denoising. Validation against ALS revisits across 19 sites and 5,087 National Forest Inventory plots shows that FORMSpoT-$Δ$ substantially outperforms existing disturbance products. In mountainous forests, where disturbances are small and spatially fragmented, FORMSpoT-$Δ$ achieves an F1-score of 0.44, representing an order of magnitude higher than existing benchmarks. By enabling tree-level monitoring of forest dynamics at national scale, FORMSpoT-$Δ$ provides a unique tool to analyze management practices, detect early signals of forest decline, and better quantify carbon losses from subtle disturbances such as thinning or selective logging. These results underscore the critical importance of sustaining very high-resolution satellite missions like SPOT and open-data initiatives such as DINAMIS for monitoring forests under climate change.
DUNIA: Pixel-Sized Embeddings via Cross-Modal Alignment for Earth Observation Applications
Feb 24, 2025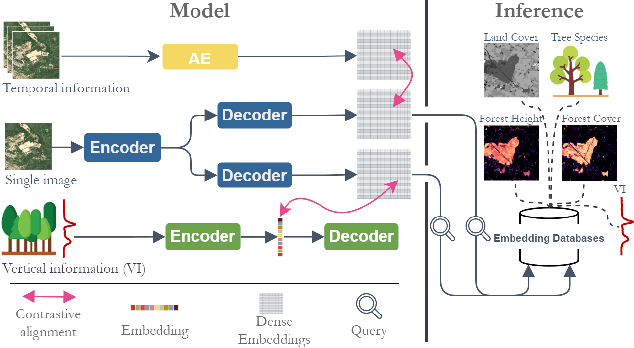
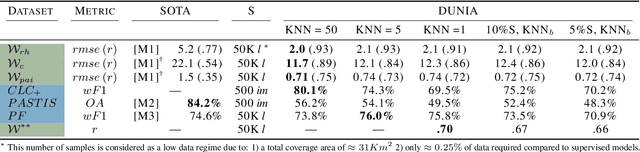
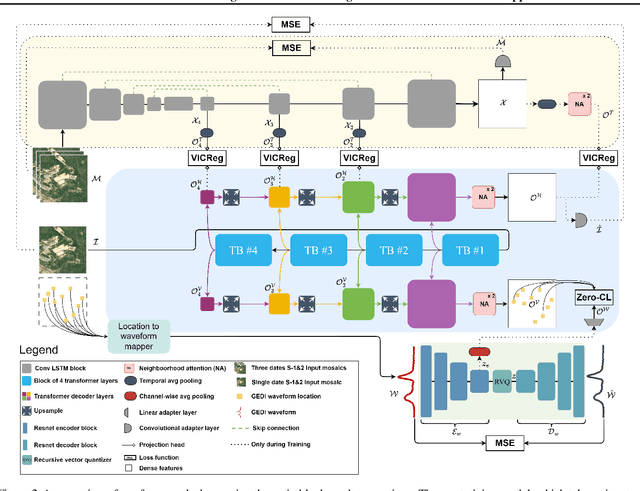
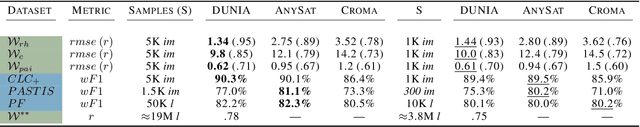
Abstract:Significant efforts have been directed towards adapting self-supervised multimodal learning for Earth observation applications. However, existing methods produce coarse patch-sized embeddings, limiting their effectiveness and integration with other modalities like LiDAR. To close this gap, we present DUNIA, an approach to learn pixel-sized embeddings through cross-modal alignment between images and full-waveform LiDAR data. As the model is trained in a contrastive manner, the embeddings can be directly leveraged in the context of a variety of environmental monitoring tasks in a zero-shot setting. In our experiments, we demonstrate the effectiveness of the embeddings for seven such tasks (canopy height mapping, fractional canopy cover, land cover mapping, tree species identification, plant area index, crop type classification, and per-pixel waveform-based vertical structure mapping). The results show that the embeddings, along with zero-shot classifiers, often outperform specialized supervised models, even in low data regimes. In the fine-tuning setting, we show strong low-shot capabilities with performances near or better than state-of-the-art on five out of six tasks.
Open-Canopy: A Country-Scale Benchmark for Canopy Height Estimation at Very High Resolution
Jul 12, 2024



Abstract:Estimating canopy height and canopy height change at meter resolution from satellite imagery has numerous applications, such as monitoring forest health, logging activities, wood resources, and carbon stocks. However, many existing forest datasets are based on commercial or closed data sources, restricting the reproducibility and evaluation of new approaches. To address this gap, we introduce Open-Canopy, the first open-access and country-scale benchmark for very high resolution (1.5 m) canopy height estimation. Covering more than 87,000 km$^2$ across France, Open-Canopy combines SPOT satellite imagery with high resolution aerial LiDAR data. We also propose Open-Canopy-$\Delta$, the first benchmark for canopy height change detection between two images taken at different years, a particularly challenging task even for recent models. To establish a robust foundation for these benchmarks, we evaluate a comprehensive list of state-of-the-art computer vision models for canopy height estimation. The dataset and associated codes can be accessed at https://github.com/fajwel/Open-Canopy.
Estimating Canopy Height at Scale
Jun 03, 2024



Abstract:We propose a framework for global-scale canopy height estimation based on satellite data. Our model leverages advanced data preprocessing techniques, resorts to a novel loss function designed to counter geolocation inaccuracies inherent in the ground-truth height measurements, and employs data from the Shuttle Radar Topography Mission to effectively filter out erroneous labels in mountainous regions, enhancing the reliability of our predictions in those areas. A comparison between predictions and ground-truth labels yields an MAE / RMSE of 2.43 / 4.73 (meters) overall and 4.45 / 6.72 (meters) for trees taller than five meters, which depicts a substantial improvement compared to existing global-scale maps. The resulting height map as well as the underlying framework will facilitate and enhance ecological analyses at a global scale, including, but not limited to, large-scale forest and biomass monitoring.
Vision Transformers, a new approach for high-resolution and large-scale mapping of canopy heights
Apr 22, 2023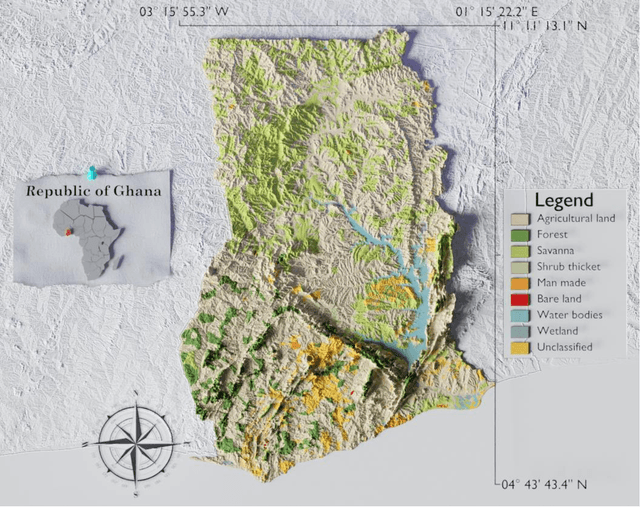
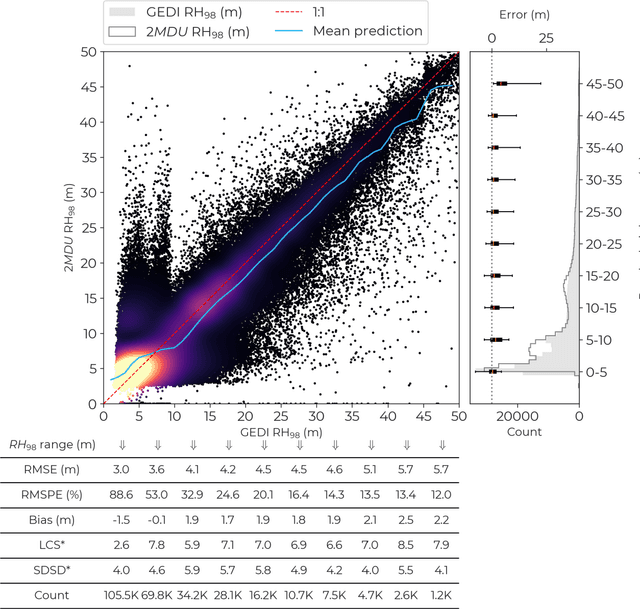
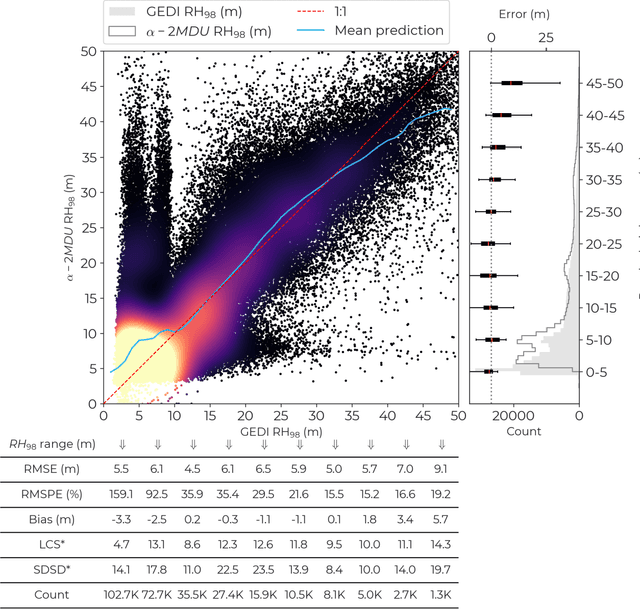
Abstract:Accurate and timely monitoring of forest canopy heights is critical for assessing forest dynamics, biodiversity, carbon sequestration as well as forest degradation and deforestation. Recent advances in deep learning techniques, coupled with the vast amount of spaceborne remote sensing data offer an unprecedented opportunity to map canopy height at high spatial and temporal resolutions. Current techniques for wall-to-wall canopy height mapping correlate remotely sensed 2D information from optical and radar sensors to the vertical structure of trees using LiDAR measurements. While studies using deep learning algorithms have shown promising performances for the accurate mapping of canopy heights, they have limitations due to the type of architectures and loss functions employed. Moreover, mapping canopy heights over tropical forests remains poorly studied, and the accurate height estimation of tall canopies is a challenge due to signal saturation from optical and radar sensors, persistent cloud covers and sometimes the limited penetration capabilities of LiDARs. Here, we map heights at 10 m resolution across the diverse landscape of Ghana with a new vision transformer (ViT) model optimized concurrently with a classification (discrete) and a regression (continuous) loss function. This model achieves better accuracy than previously used convolutional based approaches (ConvNets) optimized with only a continuous loss function. The ViT model results show that our proposed discrete/continuous loss significantly increases the sensitivity for very tall trees (i.e., > 35m), for which other approaches show saturation effects. The height maps generated by the ViT also have better ground sampling distance and better sensitivity to sparse vegetation in comparison to a convolutional model. Our ViT model has a RMSE of 3.12m in comparison to a reference dataset while the ConvNet model has a RMSE of 4.3m.
High-resolution canopy height map in the Landes forest based on GEDI, Sentinel-1, and Sentinel-2 data with a deep learning approach
Dec 20, 2022Abstract:In intensively managed forests in Europe, where forests are divided into stands of small size and may show heterogeneity within stands, a high spatial resolution (10 - 20 meters) is arguably needed to capture the differences in canopy height. In this work, we developed a deep learning model based on multi-stream remote sensing measurements to create a high-resolution canopy height map over the "Landes de Gascogne" forest in France, a large maritime pine plantation of 13,000 km$^2$ with flat terrain and intensive management. This area is characterized by even-aged and mono-specific stands, of a typical length of a few hundred meters, harvested every 35 to 50 years. Our deep learning U-Net model uses multi-band images from Sentinel-1 and Sentinel-2 with composite time averages as input to predict tree height derived from GEDI waveforms. The evaluation is performed with external validation data from forest inventory plots and a stereo 3D reconstruction model based on Skysat imagery available at specific locations. We trained seven different U-net models based on a combination of Sentinel-1 and Sentinel-2 bands to evaluate the importance of each instrument in the dominant height retrieval. The model outputs allow us to generate a 10 m resolution canopy height map of the whole "Landes de Gascogne" forest area for 2020 with a mean absolute error of 2.02 m on the Test dataset. The best predictions were obtained using all available satellite layers from Sentinel-1 and Sentinel-2 but using only one satellite source also provided good predictions. For all validation datasets in coniferous forests, our model showed better metrics than previous canopy height models available in the same region.
Fully Automated and Standardized Segmentation of Adipose Tissue Compartments by Deep Learning in Three-dimensional Whole-body MRI of Epidemiological Cohort Studies
Aug 05, 2020



Abstract:Purpose: To enable fast and reliable assessment of subcutaneous and visceral adipose tissue compartments derived from whole-body MRI. Methods: Quantification and localization of different adipose tissue compartments from whole-body MR images is of high interest to examine metabolic conditions. For correct identification and phenotyping of individuals at increased risk for metabolic diseases, a reliable automatic segmentation of adipose tissue into subcutaneous and visceral adipose tissue is required. In this work we propose a 3D convolutional neural network (DCNet) to provide a robust and objective segmentation. In this retrospective study, we collected 1000 cases (66$\pm$ 13 years; 523 women) from the Tuebingen Family Study and from the German Center for Diabetes research (TUEF/DZD), as well as 300 cases (53$\pm$ 11 years; 152 women) from the German National Cohort (NAKO) database for model training, validation, and testing with a transfer learning between the cohorts. These datasets had variable imaging sequences, imaging contrasts, receiver coil arrangements, scanners and imaging field strengths. The proposed DCNet was compared against a comparable 3D UNet segmentation in terms of sensitivity, specificity, precision, accuracy, and Dice overlap. Results: Fast (5-7seconds) and reliable adipose tissue segmentation can be obtained with high Dice overlap (0.94), sensitivity (96.6%), specificity (95.1%), precision (92.1%) and accuracy (98.4%) from 3D whole-body MR datasets (field of view coverage 450x450x2000mm${}^3$). Segmentation masks and adipose tissue profiles are automatically reported back to the referring physician. Conclusion: Automatic adipose tissue segmentation is feasible in 3D whole-body MR data sets and is generalizable to different epidemiological cohort studies with the proposed DCNet.
A Machine-learning framework for automatic reference-free quality assessment in MRI
Jul 18, 2018



Abstract:Magnetic resonance (MR) imaging offers a wide variety of imaging techniques. A large amount of data is created per examination which needs to be checked for sufficient quality in order to derive a meaningful diagnosis. This is a manual process and therefore time- and cost-intensive. Any imaging artifacts originating from scanner hardware, signal processing or induced by the patient may reduce the image quality and complicate the diagnosis or any image post-processing. Therefore, the assessment or the ensurance of sufficient image quality in an automated manner is of high interest. Usually no reference image is available or difficult to define. Therefore, classical reference-based approaches are not applicable. Model observers mimicking the human observers (HO) can assist in this task. Thus, we propose a new machine-learning-based reference-free MR image quality assessment framework which is trained on HO-derived labels to assess MR image quality immediately after each acquisition. We include the concept of active learning and present an efficient blinded reading platform to reduce the effort in the HO labeling procedure. Derived image features and the applied classifiers (support-vector-machine, deep neural network) are investigated for a cohort of 250 patients. The MR image quality assessment framework can achieve a high test accuracy of 93.7$\%$ for estimating quality classes on a 5-point Likert-scale. The proposed MR image quality assessment framework is able to provide an accurate and efficient quality estimation which can be used as a prospective quality assurance including automatic acquisition adaptation or guided MR scanner operation, and/or as a retrospective quality assessment including support of diagnostic decisions or quality control in cohort studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge