Konstantin Nikolaou
Requirements for Quality Assurance of AI Models for Early Detection of Lung Cancer
Feb 24, 2025Abstract:Lung cancer is the second most common cancer and the leading cause of cancer-related deaths worldwide. Survival largely depends on tumor stage at diagnosis, and early detection with low-dose CT can significantly reduce mortality in high-risk patients. AI can improve the detection, measurement, and characterization of pulmonary nodules while reducing assessment time. However, the training data, functionality, and performance of available AI systems vary considerably, complicating software selection and regulatory evaluation. Manufacturers must specify intended use and provide test statistics, but they can choose their training and test data, limiting standardization and comparability. Under the EU AI Act, consistent quality assurance is required for AI-based nodule detection, measurement, and characterization. This position paper proposes systematic quality assurance grounded in a validated reference dataset, including real screening cases plus phantom data to verify volume and growth rate measurements. Regular updates shall reflect demographic shifts and technological advances, ensuring ongoing relevance. Consequently, ongoing AI quality assurance is vital. Regulatory challenges are also adressed. While the MDR and the EU AI Act set baseline requirements, they do not adequately address self-learning algorithms or their updates. A standardized, transparent quality assessment - based on sensitivity, specificity, and volumetric accuracy - enables an objective evaluation of each AI solution's strengths and weaknesses. Establishing clear testing criteria and systematically using updated reference data lay the groundwork for comparable performance metrics, informing tenders, guidelines, and recommendations.
Collective variables of neural networks: empirical time evolution and scaling laws
Oct 09, 2024
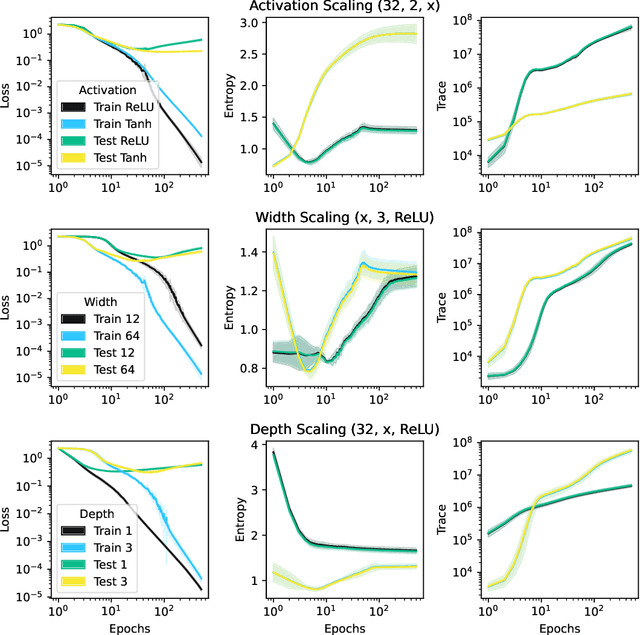
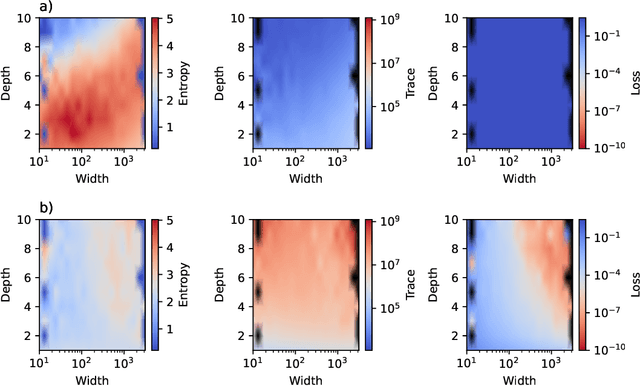
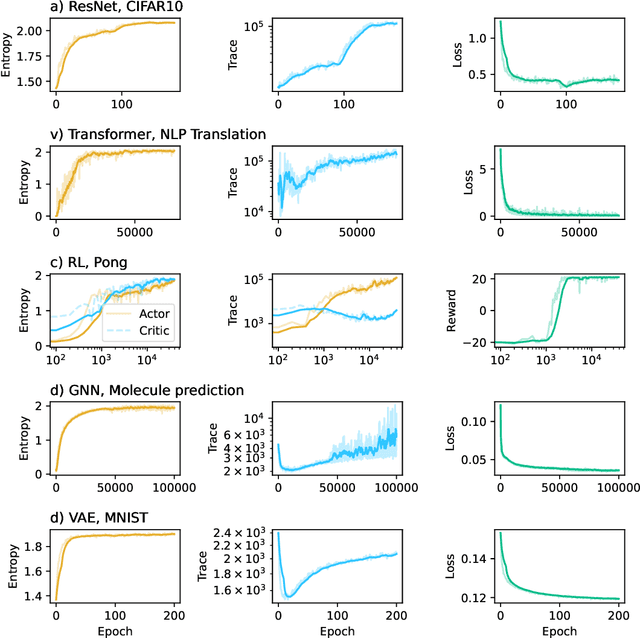
Abstract:This work presents a novel means for understanding learning dynamics and scaling relations in neural networks. We show that certain measures on the spectrum of the empirical neural tangent kernel, specifically entropy and trace, yield insight into the representations learned by a neural network and how these can be improved through architecture scaling. These results are demonstrated first on test cases before being shown on more complex networks, including transformers, auto-encoders, graph neural networks, and reinforcement learning studies. In testing on a wide range of architectures, we highlight the universal nature of training dynamics and further discuss how it can be used to understand the mechanisms behind learning in neural networks. We identify two such dominant mechanisms present throughout machine learning training. The first, information compression, is seen through a reduction in the entropy of the NTK spectrum during training, and occurs predominantly in small neural networks. The second, coined structure formation, is seen through an increasing entropy and thus, the creation of structure in the neural network representations beyond the prior established by the network at initialization. Due to the ubiquity of the latter in deep neural network architectures and its flexibility in the creation of feature-rich representations, we argue that this form of evolution of the network's entropy be considered the onset of a deep learning regime.
SwarmRL: Building the Future of Smart Active Systems
Apr 25, 2024
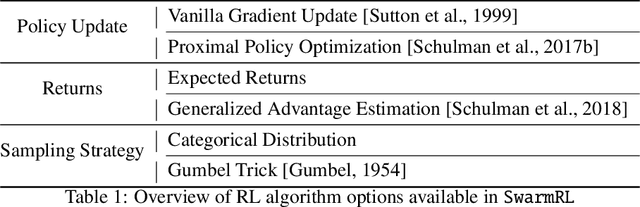


Abstract:This work introduces SwarmRL, a Python package designed to study intelligent active particles. SwarmRL provides an easy-to-use interface for developing models to control microscopic colloids using classical control and deep reinforcement learning approaches. These models may be deployed in simulations or real-world environments under a common framework. We explain the structure of the software and its key features and demonstrate how it can be used to accelerate research. With SwarmRL, we aim to streamline research into micro-robotic control while bridging the gap between experimental and simulation-driven sciences. SwarmRL is available open-source on GitHub at https://github.com/SwarmRL/SwarmRL.
Towards a Phenomenological Understanding of Neural Networks: Data
May 01, 2023Abstract:A theory of neural networks (NNs) built upon collective variables would provide scientists with the tools to better understand the learning process at every stage. In this work, we introduce two such variables, the entropy and the trace of the empirical neural tangent kernel (NTK) built on the training data passed to the model. We empirically analyze the NN performance in the context of these variables and find that there exists correlation between the starting entropy, the trace of the NTK, and the generalization of the model computed after training is complete. This framework is then applied to the problem of optimal data selection for the training of NNs. To this end, random network distillation (RND) is used as a means of selecting training data which is then compared with random selection of data. It is shown that not only does RND select data-sets capable of outperforming random selection, but that the collective variables associated with the RND data-sets are larger than those of the randomly selected sets. The results of this investigation provide a stable ground from which the selection of data for NN training can be driven by this phenomenological framework.
Fully Automated and Standardized Segmentation of Adipose Tissue Compartments by Deep Learning in Three-dimensional Whole-body MRI of Epidemiological Cohort Studies
Aug 05, 2020



Abstract:Purpose: To enable fast and reliable assessment of subcutaneous and visceral adipose tissue compartments derived from whole-body MRI. Methods: Quantification and localization of different adipose tissue compartments from whole-body MR images is of high interest to examine metabolic conditions. For correct identification and phenotyping of individuals at increased risk for metabolic diseases, a reliable automatic segmentation of adipose tissue into subcutaneous and visceral adipose tissue is required. In this work we propose a 3D convolutional neural network (DCNet) to provide a robust and objective segmentation. In this retrospective study, we collected 1000 cases (66$\pm$ 13 years; 523 women) from the Tuebingen Family Study and from the German Center for Diabetes research (TUEF/DZD), as well as 300 cases (53$\pm$ 11 years; 152 women) from the German National Cohort (NAKO) database for model training, validation, and testing with a transfer learning between the cohorts. These datasets had variable imaging sequences, imaging contrasts, receiver coil arrangements, scanners and imaging field strengths. The proposed DCNet was compared against a comparable 3D UNet segmentation in terms of sensitivity, specificity, precision, accuracy, and Dice overlap. Results: Fast (5-7seconds) and reliable adipose tissue segmentation can be obtained with high Dice overlap (0.94), sensitivity (96.6%), specificity (95.1%), precision (92.1%) and accuracy (98.4%) from 3D whole-body MR datasets (field of view coverage 450x450x2000mm${}^3$). Segmentation masks and adipose tissue profiles are automatically reported back to the referring physician. Conclusion: Automatic adipose tissue segmentation is feasible in 3D whole-body MR data sets and is generalizable to different epidemiological cohort studies with the proposed DCNet.
Retrospective correction of Rigid and Non-Rigid MR motion artifacts using GANs
Oct 06, 2018


Abstract:Motion artifacts are a primary source of magnetic resonance (MR) image quality deterioration with strong repercussions on diagnostic performance. Currently, MR motion correction is carried out either prospectively, with the help of motion tracking systems, or retrospectively by mainly utilizing computationally expensive iterative algorithms. In this paper, we utilize a new adversarial framework, titled MedGAN, for the joint retrospective correction of rigid and non-rigid motion artifacts in different body regions and without the need for a reference image. MedGAN utilizes a unique combination of non-adversarial losses and a new generator architecture to capture the textures and fine-detailed structures of the desired artifact-free MR images. Quantitative and qualitative comparisons with other adversarial techniques have illustrated the proposed model performance.
A Machine-learning framework for automatic reference-free quality assessment in MRI
Jul 18, 2018



Abstract:Magnetic resonance (MR) imaging offers a wide variety of imaging techniques. A large amount of data is created per examination which needs to be checked for sufficient quality in order to derive a meaningful diagnosis. This is a manual process and therefore time- and cost-intensive. Any imaging artifacts originating from scanner hardware, signal processing or induced by the patient may reduce the image quality and complicate the diagnosis or any image post-processing. Therefore, the assessment or the ensurance of sufficient image quality in an automated manner is of high interest. Usually no reference image is available or difficult to define. Therefore, classical reference-based approaches are not applicable. Model observers mimicking the human observers (HO) can assist in this task. Thus, we propose a new machine-learning-based reference-free MR image quality assessment framework which is trained on HO-derived labels to assess MR image quality immediately after each acquisition. We include the concept of active learning and present an efficient blinded reading platform to reduce the effort in the HO labeling procedure. Derived image features and the applied classifiers (support-vector-machine, deep neural network) are investigated for a cohort of 250 patients. The MR image quality assessment framework can achieve a high test accuracy of 93.7$\%$ for estimating quality classes on a 5-point Likert-scale. The proposed MR image quality assessment framework is able to provide an accurate and efficient quality estimation which can be used as a prospective quality assurance including automatic acquisition adaptation or guided MR scanner operation, and/or as a retrospective quality assessment including support of diagnostic decisions or quality control in cohort studies.
MedGAN: Medical Image Translation using GANs
Jun 17, 2018



Abstract:Image-to-image translation is considered a next frontier in the field of medical image analysis, with numerous potential applications. However, recent advances in this field offer individualized solutions by utilizing specialized architectures which are task specific or by suffering from limited capacities and thus requiring refinement through non end-to-end training. In this paper, we propose a novel general purpose framework for medical image-to-image translation, titled MedGAN, which operates in an end-to-end manner on the image level. MedGAN builds upon recent advances in the field of generative adversarial networks(GANs) by combining the adversarial framework with a unique combination of non-adversarial losses which captures the high and low frequency components of the desired target modality. Namely, we utilize a discriminator network as a trainable feature extractor which penalizes the discrepancy between the translated medical images and the desired modalities in the pixel and perceptual sense. Moreover, style-transfer losses are utilized to match the textures and fine-structures of the desired target images to the outputs. Additionally, we present a novel generator architecture, titled CasNet, which enhances the sharpness of the translated medical outputs through progressive refinement via encoder decoder pairs. To demonstrate the effectiveness of our approach, we apply MedGAN on three novel and challenging applications: PET-CT translation, correction of MR motion artefacts and PET image denoising. Qualitative and quantitative comparisons with state-of-the-art techniques have emphasized the superior performance of the proposed framework. MedGAN can be directly applied as a general framework for future medical translation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge