Christian Holm
Deep Learning-Driven Peptide Classification in Biological Nanopores
Sep 17, 2025



Abstract:A device capable of performing real time classification of proteins in a clinical setting would allow for inexpensive and rapid disease diagnosis. One such candidate for this technology are nanopore devices. These devices work by measuring a current signal that arises when a protein or peptide enters a nanometer-length-scale pore. Should this current be uniquely related to the structure of the peptide and its interactions with the pore, the signals can be used to perform identification. While such a method would allow for real time identification of peptides and proteins in a clinical setting, to date, the complexities of these signals limit their accuracy. In this work, we tackle the issue of classification by converting the current signals into scaleogram images via wavelet transforms, capturing amplitude, frequency, and time information in a modality well-suited to machine learning algorithms. When tested on 42 peptides, our method achieved a classification accuracy of ~$81\,\%$, setting a new state-of-the-art in the field and taking a step toward practical peptide/protein diagnostics at the point of care. In addition, we demonstrate model transfer techniques that will be critical when deploying these models into real hardware, paving the way to a new method for real-time disease diagnosis.
The interplay of robustness and generalization in quantum machine learning
Jun 10, 2025Abstract:While adversarial robustness and generalization have individually received substantial attention in the recent literature on quantum machine learning, their interplay is much less explored. In this chapter, we address this interplay for variational quantum models, which were recently proposed as function approximators in supervised learning. We discuss recent results quantifying both robustness and generalization via Lipschitz bounds, which explicitly depend on model parameters. Thus, they give rise to a regularization-based training approach for robust and generalizable quantum models, highlighting the importance of trainable data encoding strategies. The practical implications of the theoretical results are demonstrated with an application to time series analysis.
Collective variables of neural networks: empirical time evolution and scaling laws
Oct 09, 2024
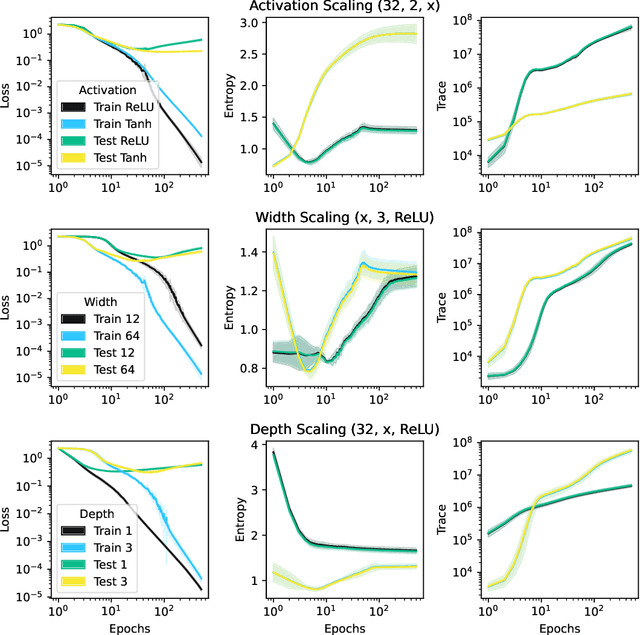
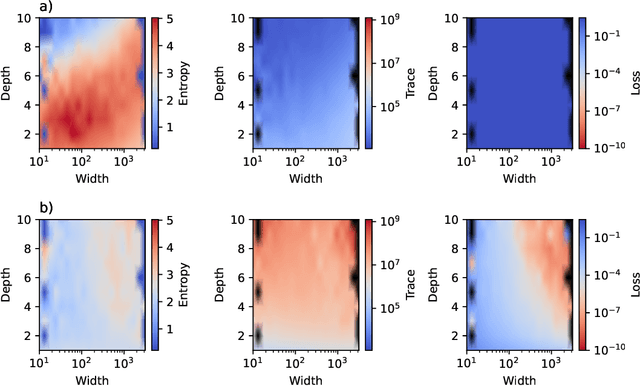
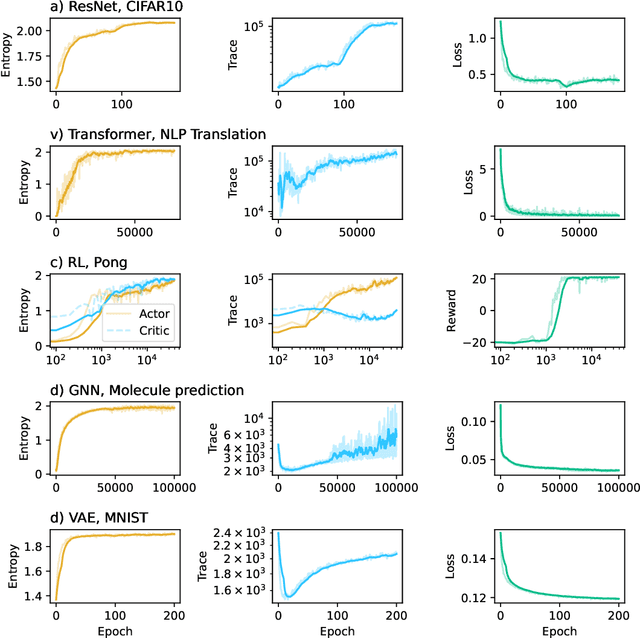
Abstract:This work presents a novel means for understanding learning dynamics and scaling relations in neural networks. We show that certain measures on the spectrum of the empirical neural tangent kernel, specifically entropy and trace, yield insight into the representations learned by a neural network and how these can be improved through architecture scaling. These results are demonstrated first on test cases before being shown on more complex networks, including transformers, auto-encoders, graph neural networks, and reinforcement learning studies. In testing on a wide range of architectures, we highlight the universal nature of training dynamics and further discuss how it can be used to understand the mechanisms behind learning in neural networks. We identify two such dominant mechanisms present throughout machine learning training. The first, information compression, is seen through a reduction in the entropy of the NTK spectrum during training, and occurs predominantly in small neural networks. The second, coined structure formation, is seen through an increasing entropy and thus, the creation of structure in the neural network representations beyond the prior established by the network at initialization. Due to the ubiquity of the latter in deep neural network architectures and its flexibility in the creation of feature-rich representations, we argue that this form of evolution of the network's entropy be considered the onset of a deep learning regime.
SwarmRL: Building the Future of Smart Active Systems
Apr 25, 2024
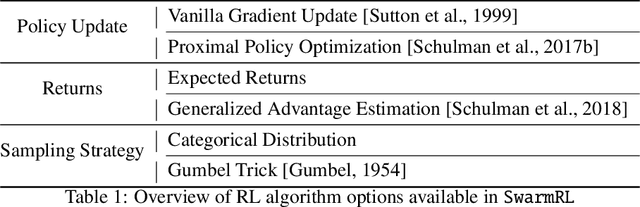


Abstract:This work introduces SwarmRL, a Python package designed to study intelligent active particles. SwarmRL provides an easy-to-use interface for developing models to control microscopic colloids using classical control and deep reinforcement learning approaches. These models may be deployed in simulations or real-world environments under a common framework. We explain the structure of the software and its key features and demonstrate how it can be used to accelerate research. With SwarmRL, we aim to streamline research into micro-robotic control while bridging the gap between experimental and simulation-driven sciences. SwarmRL is available open-source on GitHub at https://github.com/SwarmRL/SwarmRL.
Emergence of Chemotactic Strategies with Multi-Agent Reinforcement Learning
Apr 02, 2024



Abstract:Reinforcement learning (RL) is a flexible and efficient method for programming micro-robots in complex environments. Here we investigate whether reinforcement learning can provide insights into biological systems when trained to perform chemotaxis. Namely, whether we can learn about how intelligent agents process given information in order to swim towards a target. We run simulations covering a range of agent shapes, sizes, and swim speeds to determine if the physical constraints on biological swimmers, namely Brownian motion, lead to regions where reinforcement learners' training fails. We find that the RL agents can perform chemotaxis as soon as it is physically possible and, in some cases, even before the active swimming overpowers the stochastic environment. We study the efficiency of the emergent policy and identify convergence in agent size and swim speeds. Finally, we study the strategy adopted by the reinforcement learning algorithm to explain how the agents perform their tasks. To this end, we identify three emerging dominant strategies and several rare approaches taken. These strategies, whilst producing almost identical trajectories in simulation, are distinct and give insight into the possible mechanisms behind which biological agents explore their environment and respond to changing conditions.
Zero Shot Molecular Generation via Similarity Kernels
Feb 13, 2024



Abstract:Generative modelling aims to accelerate the discovery of novel chemicals by directly proposing structures with desirable properties. Recently, score-based, or diffusion, generative models have significantly outperformed previous approaches. Key to their success is the close relationship between the score and physical force, allowing the use of powerful equivariant neural networks. However, the behaviour of the learnt score is not yet well understood. Here, we analyse the score by training an energy-based diffusion model for molecular generation. We find that during the generation the score resembles a restorative potential initially and a quantum-mechanical force at the end. In between the two endpoints, it exhibits special properties that enable the building of large molecules. Using insights from the trained model, we present Similarity-based Molecular Generation (SiMGen), a new method for zero shot molecular generation. SiMGen combines a time-dependent similarity kernel with descriptors from a pretrained machine learning force field to generate molecules without any further training. Our approach allows full control over the molecular shape through point cloud priors and supports conditional generation. We also release an interactive web tool that allows users to generate structures with SiMGen online (https://zndraw.icp.uni-stuttgart.de).
ZnTrack -- Data as Code
Jan 19, 2024Abstract:The past decade has seen tremendous breakthroughs in computation and there is no indication that this will slow any time soon. Machine learning, large-scale computing resources, and increased industry focus have resulted in rising investments in computer-driven solutions for data management, simulations, and model generation. However, with this growth in computation has come an even larger expansion of data and with it, complexity in data storage, sharing, and tracking. In this work, we introduce ZnTrack, a Python-driven data versioning tool. ZnTrack builds upon established version control systems to provide a user-friendly and easy-to-use interface for tracking parameters in experiments, designing workflows, and storing and sharing data. From this ability to reduce large datasets to a simple Python script emerges the concept of Data as Code, a core component of the work presented here and an undoubtedly important concept as the age of computation continues to evolve. ZnTrack offers an open-source, FAIR data compatible Python package to enable users to harness these concepts of the future.
Training robust and generalizable quantum models
Nov 20, 2023



Abstract:Adversarial robustness and generalization are both crucial properties of reliable machine learning models. In this paper, we study these properties in the context of quantum machine learning based on Lipschitz bounds. We derive tailored, parameter-dependent Lipschitz bounds for quantum models with trainable encoding, showing that the norm of the data encoding has a crucial impact on the robustness against perturbations in the input data. Further, we derive a bound on the generalization error which explicitly depends on the parameters of the data encoding. Our theoretical findings give rise to a practical strategy for training robust and generalizable quantum models by regularizing the Lipschitz bound in the cost. Further, we show that, for fixed and non-trainable encodings as frequently employed in quantum machine learning, the Lipschitz bound cannot be influenced by tuning the parameters. Thus, trainable encodings are crucial for systematically adapting robustness and generalization during training. With numerical results, we demonstrate that, indeed, Lipschitz bound regularization leads to substantially more robust and generalizable quantum models.
Environmental effects on emergent strategy in micro-scale multi-agent reinforcement learning
Jul 03, 2023



Abstract:Multi-Agent Reinforcement Learning (MARL) is a promising candidate for realizing efficient control of microscopic particles, of which micro-robots are a subset. However, the microscopic particles' environment presents unique challenges, such as Brownian motion at sufficiently small length-scales. In this work, we explore the role of temperature in the emergence and efficacy of strategies in MARL systems using particle-based Langevin molecular dynamics simulations as a realistic representation of micro-scale environments. To this end, we perform experiments on two different multi-agent tasks in microscopic environments at different temperatures, detecting the source of a concentration gradient and rotation of a rod. We find that at higher temperatures, the RL agents identify new strategies for achieving these tasks, highlighting the importance of understanding this regime and providing insight into optimal training strategies for bridging the generalization gap between simulation and reality. We also introduce a novel Python package for studying microscopic agents using reinforcement learning (RL) to accompany our results.
Towards a Phenomenological Understanding of Neural Networks: Data
May 01, 2023Abstract:A theory of neural networks (NNs) built upon collective variables would provide scientists with the tools to better understand the learning process at every stage. In this work, we introduce two such variables, the entropy and the trace of the empirical neural tangent kernel (NTK) built on the training data passed to the model. We empirically analyze the NN performance in the context of these variables and find that there exists correlation between the starting entropy, the trace of the NTK, and the generalization of the model computed after training is complete. This framework is then applied to the problem of optimal data selection for the training of NNs. To this end, random network distillation (RND) is used as a means of selecting training data which is then compared with random selection of data. It is shown that not only does RND select data-sets capable of outperforming random selection, but that the collective variables associated with the RND data-sets are larger than those of the randomly selected sets. The results of this investigation provide a stable ground from which the selection of data for NN training can be driven by this phenomenological framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge