Daniel Fink
Cornell Lab of Ornithology, Cornell University, USA
LabelKAN -- Kolmogorov-Arnold Networks for Inter-Label Learning: Avian Community Learning
Jan 23, 2026Abstract:Global biodiversity loss is accelerating, prompting international efforts such as the Kunming-Montreal Global Biodiversity Framework (GBF) and the United Nations Sustainable Development Goals to direct resources toward halting species declines. A key challenge in achieving this goal is having access to robust methodologies to understand where species occur and how they relate to each other within broader ecological communities. Recent deep learning-based advances in joint species distribution modeling have shown improved predictive performance, but effectively incorporating community-level learning, taking into account species-species relationships in addition to species-environment relationships, remains an outstanding challenge. We introduce LabelKAN, a novel framework based on Kolmogorov-Arnold Networks (KANs) to learn inter-label connections from predictions of each label. When modeling avian species distributions, LabelKAN achieves substantial gains in predictive performance across the vast majority of species. In particular, our method demonstrates strong improvements for rare and difficult-to-predict species, which are often the most important when setting biodiversity targets under frameworks like GBF. These performance gains also translate to more confident predictions of the species spatial patterns as well as more confident predictions of community structure. We illustrate how the LabelKAN leads to qualitative and quantitative improvements with a focused application on the Great Blue Heron, an emblematic species in freshwater ecosystems that has experienced significant population declines across the United States in recent years. Using the LabelKAN framework, we are able to identify communities and species in New York that will be most sensitive to further declines in Great Blue Heron populations.
Training robust and generalizable quantum models
Nov 20, 2023



Abstract:Adversarial robustness and generalization are both crucial properties of reliable machine learning models. In this paper, we study these properties in the context of quantum machine learning based on Lipschitz bounds. We derive tailored, parameter-dependent Lipschitz bounds for quantum models with trainable encoding, showing that the norm of the data encoding has a crucial impact on the robustness against perturbations in the input data. Further, we derive a bound on the generalization error which explicitly depends on the parameters of the data encoding. Our theoretical findings give rise to a practical strategy for training robust and generalizable quantum models by regularizing the Lipschitz bound in the cost. Further, we show that, for fixed and non-trainable encodings as frequently employed in quantum machine learning, the Lipschitz bound cannot be influenced by tuning the parameters. Thus, trainable encodings are crucial for systematically adapting robustness and generalization during training. With numerical results, we demonstrate that, indeed, Lipschitz bound regularization leads to substantially more robust and generalizable quantum models.
A Double Machine Learning Trend Model for Citizen Science Data
Oct 27, 2022Abstract:1. Citizen and community-science (CS) datasets have great potential for estimating interannual patterns of population change given the large volumes of data collected globally every year. Yet, the flexible protocols that enable many CS projects to collect large volumes of data typically lack the structure necessary to keep consistent sampling across years. This leads to interannual confounding, as changes to the observation process over time are confounded with changes in species population sizes. 2. Here we describe a novel modeling approach designed to estimate species population trends while controlling for the interannual confounding common in citizen science data. The approach is based on Double Machine Learning, a statistical framework that uses machine learning methods to estimate population change and the propensity scores used to adjust for confounding discovered in the data. Additionally, we develop a simulation method to identify and adjust for residual confounding missed by the propensity scores. Using this new method, we can produce spatially detailed trend estimates from citizen science data. 3. To illustrate the approach, we estimated species trends using data from the CS project eBird. We used a simulation study to assess the ability of the method to estimate spatially varying trends in the face of real-world confounding. Results showed that the trend estimates distinguished between spatially constant and spatially varying trends at a 27km resolution. There were low error rates on the estimated direction of population change (increasing/decreasing) and high correlations on the estimated magnitude. 4. The ability to estimate spatially explicit trends while accounting for confounding in citizen science data has the potential to fill important information gaps, helping to estimate population trends for species, regions, or seasons without rigorous monitoring data.
HOT-VAE: Learning High-Order Label Correlation for Multi-Label Classification via Attention-Based Variational Autoencoders
Mar 09, 2021

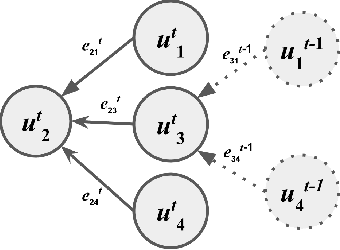

Abstract:Understanding how environmental characteristics affect bio-diversity patterns, from individual species to communities of species, is critical for mitigating effects of global change. A central goal for conservation planning and monitoring is the ability to accurately predict the occurrence of species communities and how these communities change over space and time. This in turn leads to a challenging and long-standing problem in the field of computer science - how to perform ac-curate multi-label classification with hundreds of labels? The key challenge of this problem is its exponential-sized output space with regards to the number of labels to be predicted.Therefore, it is essential to facilitate the learning process by exploiting correlations (or dependency) among labels. Previous methods mostly focus on modelling the correlation on label pairs; however, complex relations between real-world objects often go beyond second order. In this paper, we pro-pose a novel framework for multi-label classification, High-order Tie-in Variational Autoencoder (HOT-VAE), which per-forms adaptive high-order label correlation learning. We experimentally verify that our model outperforms the existing state-of-the-art approaches on a bird distribution dataset on both conventional F1 scores and a variety of ecological metrics. To show our method is general, we also perform empirical analysis on seven other public real-world datasets in several application domains, and Hot-VAE exhibits superior performance to previous methods.
Enhanced Optimization with Composite Objectives and Novelty Selection
Jul 05, 2018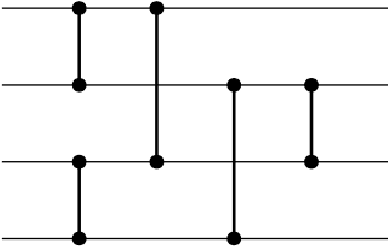
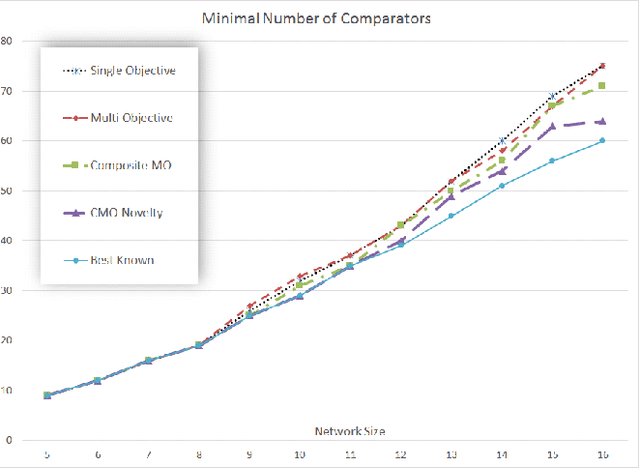
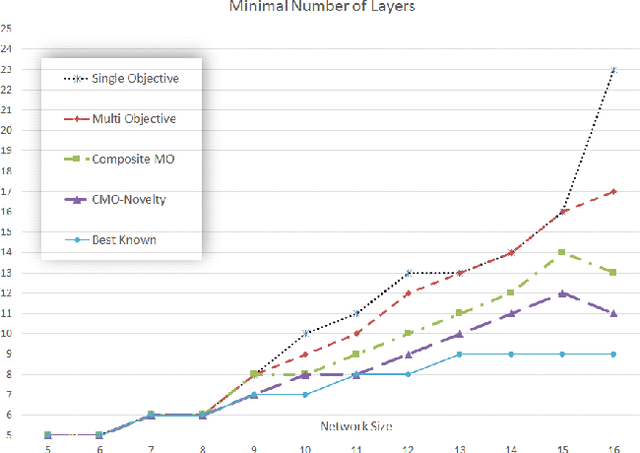
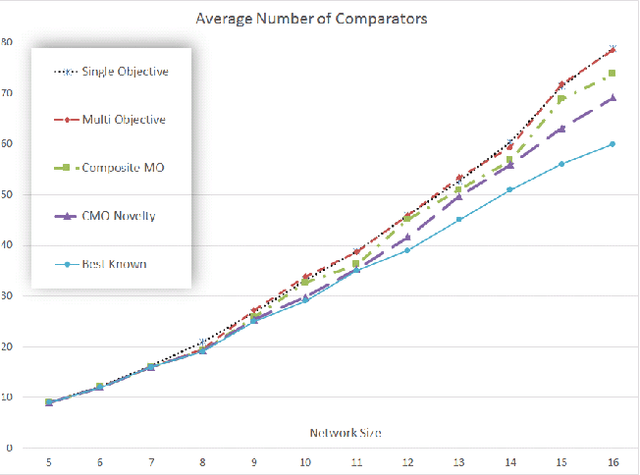
Abstract:An important benefit of multi-objective search is that it maintains a diverse population of candidates, which helps in deceptive problems in particular. Not all diversity is useful, however: candidates that optimize only one objective while ignoring others are rarely helpful. This paper proposes a solution: The original objectives are replaced by their linear combinations, thus focusing the search on the most useful tradeoffs between objectives. To compensate for the loss of diversity, this transformation is accompanied by a selection mechanism that favors novelty. In the highly deceptive problem of discovering minimal sorting networks, this approach finds better solutions, and finds them faster and more consistently than standard methods. It is therefore a promising approach to solving deceptive problems through multi-objective optimization.
Deep Multi-Species Embedding
Feb 21, 2017
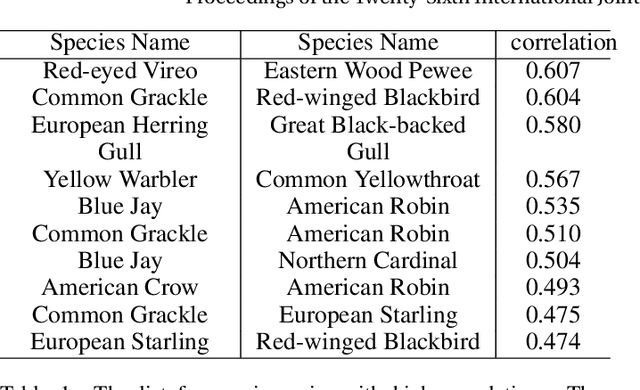

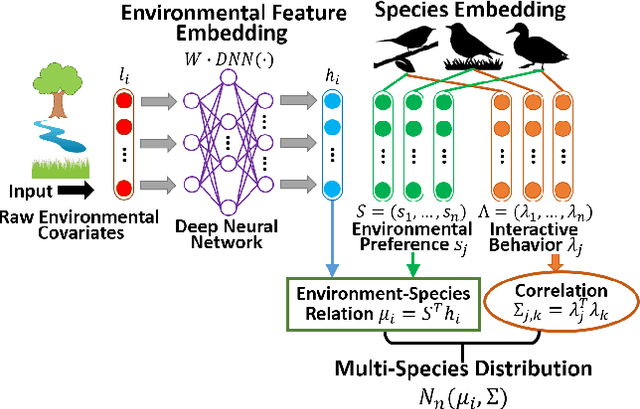
Abstract:Understanding how species are distributed across landscapes over time is a fundamental question in biodiversity research. Unfortunately, most species distribution models only target a single species at a time, despite strong ecological evidence that species are not independently distributed. We propose Deep Multi-Species Embedding (DMSE), which jointly embeds vectors corresponding to multiple species as well as vectors representing environmental covariates into a common high-dimensional feature space via a deep neural network. Applied to bird observational data from the citizen science project \textit{eBird}, we demonstrate how the DMSE model discovers inter-species relationships to outperform single-species distribution models (random forests and SVMs) as well as competing multi-label models. Additionally, we demonstrate the benefit of using a deep neural network to extract features within the embedding and show how they improve the predictive performance of species distribution modelling. An important domain contribution of the DMSE model is the ability to discover and describe species interactions while simultaneously learning the shared habitat preferences among species. As an additional contribution, we provide a graphical embedding of hundreds of bird species in the Northeast US.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge