Steve Kelling
Cornell Lab of Ornithology, Cornell University, USA
A Double Machine Learning Trend Model for Citizen Science Data
Oct 27, 2022Abstract:1. Citizen and community-science (CS) datasets have great potential for estimating interannual patterns of population change given the large volumes of data collected globally every year. Yet, the flexible protocols that enable many CS projects to collect large volumes of data typically lack the structure necessary to keep consistent sampling across years. This leads to interannual confounding, as changes to the observation process over time are confounded with changes in species population sizes. 2. Here we describe a novel modeling approach designed to estimate species population trends while controlling for the interannual confounding common in citizen science data. The approach is based on Double Machine Learning, a statistical framework that uses machine learning methods to estimate population change and the propensity scores used to adjust for confounding discovered in the data. Additionally, we develop a simulation method to identify and adjust for residual confounding missed by the propensity scores. Using this new method, we can produce spatially detailed trend estimates from citizen science data. 3. To illustrate the approach, we estimated species trends using data from the CS project eBird. We used a simulation study to assess the ability of the method to estimate spatially varying trends in the face of real-world confounding. Results showed that the trend estimates distinguished between spatially constant and spatially varying trends at a 27km resolution. There were low error rates on the estimated direction of population change (increasing/decreasing) and high correlations on the estimated magnitude. 4. The ability to estimate spatially explicit trends while accounting for confounding in citizen science data has the potential to fill important information gaps, helping to estimate population trends for species, regions, or seasons without rigorous monitoring data.
Robust sound event detection in bioacoustic sensor networks
May 20, 2019

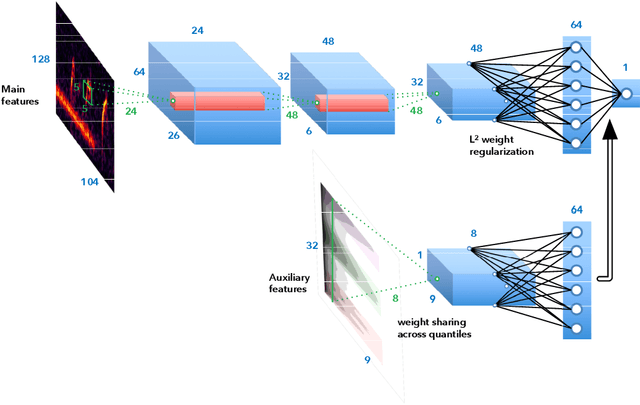
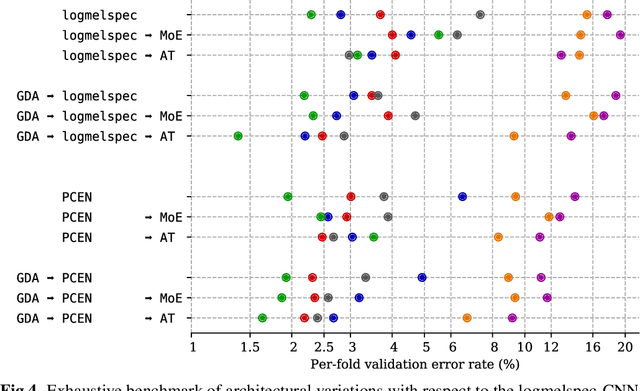
Abstract:Bioacoustic sensors, sometimes known as autonomous recording units (ARUs), can record sounds of wildlife over long periods of time in scalable and minimally invasive ways. Deriving per-species abundance estimates from these sensors requires detection, classification, and quantification of animal vocalizations as individual acoustic events. Yet, variability in ambient noise, both over time and across sensors, hinders the reliability of current automated systems for sound event detection (SED), such as convolutional neural networks (CNN) in the time-frequency domain. In this article, we develop, benchmark, and combine several machine listening techniques to improve the generalizability of SED models across heterogeneous acoustic environments. As a case study, we consider the problem of detecting avian flight calls from a ten-hour recording of nocturnal bird migration, recorded by a network of six ARUs in the presence of heterogeneous background noise. Starting from a CNN yielding state-of-the-art accuracy on this task, we introduce two noise adaptation techniques, respectively integrating short-term (60-millisecond) and long-term (30-minute) context. First, we apply per-channel energy normalization (PCEN) in the time-frequency domain, which applies short-term automatic gain control to every subband in the mel-frequency spectrogram. Secondly, we replace the last dense layer in the network by a context-adaptive neural network (CA-NN) layer, i.e. an affine layer whose weights are dynamically adapted at prediction time by an auxiliary network taking long-term summary statistics of spectrotemporal features as input. We show that both techniques are helpful and complementary. [...] We release a pre-trained version of our best performing system under the name of BirdVoxDetect, a ready-to-use detector of avian flight calls in field recordings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge