Mario Geiger
Optimizing Data Distribution and Kernel Performance for Efficient Training of Chemistry Foundation Models: A Case Study with MACE
Apr 14, 2025



Abstract:Chemistry Foundation Models (CFMs) that leverage Graph Neural Networks (GNNs) operating on 3D molecular graph structures are becoming indispensable tools for computational chemists and materials scientists. These models facilitate the understanding of matter and the discovery of new molecules and materials. In contrast to GNNs operating on a large homogeneous graphs, GNNs used by CFMs process a large number of geometric graphs of varying sizes, requiring different optimization strategies than those developed for large homogeneous GNNs. This paper presents optimizations for two critical phases of CFM training: data distribution and model training, targeting MACE - a state-of-the-art CFM. We address the challenge of load balancing in data distribution by formulating it as a multi-objective bin packing problem. We propose an iterative algorithm that provides a highly effective, fast, and practical solution, ensuring efficient data distribution. For the training phase, we identify symmetric tensor contraction as the key computational kernel in MACE and optimize this kernel to improve the overall performance. Our combined approach of balanced data distribution and kernel optimization significantly enhances the training process of MACE. Experimental results demonstrate a substantial speedup, reducing per-epoch execution time for training from 12 to 2 minutes on 740 GPUs with a 2.6M sample dataset.
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024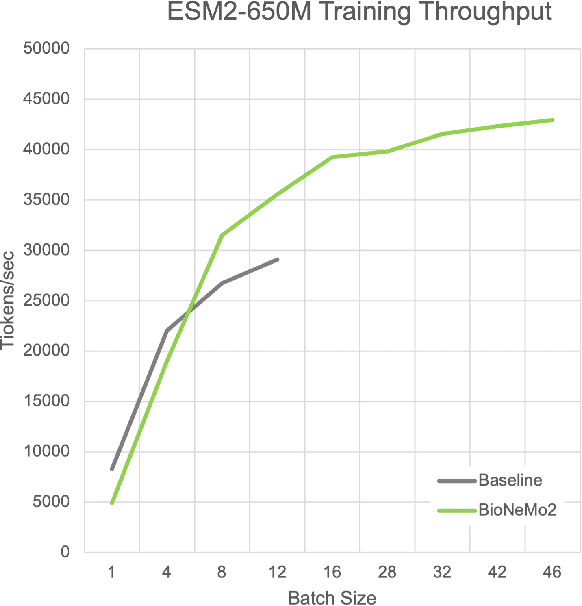
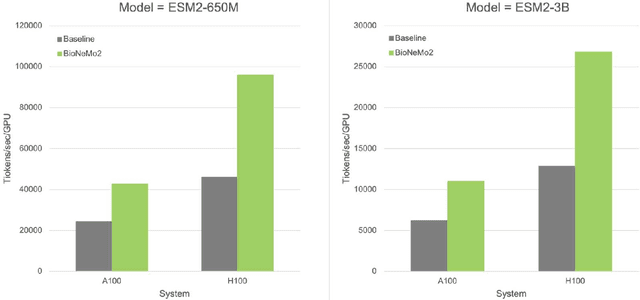
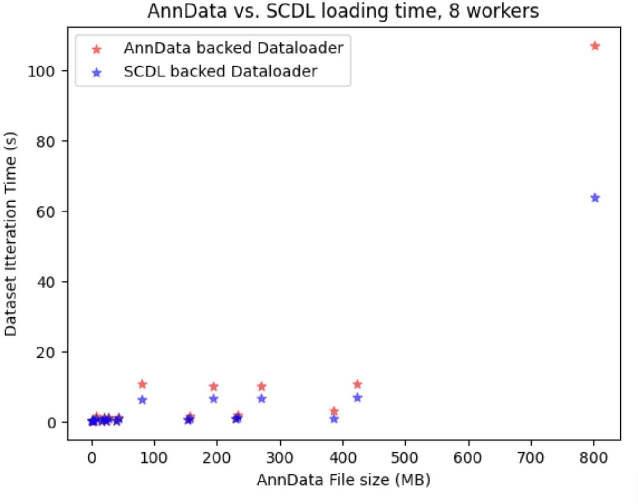
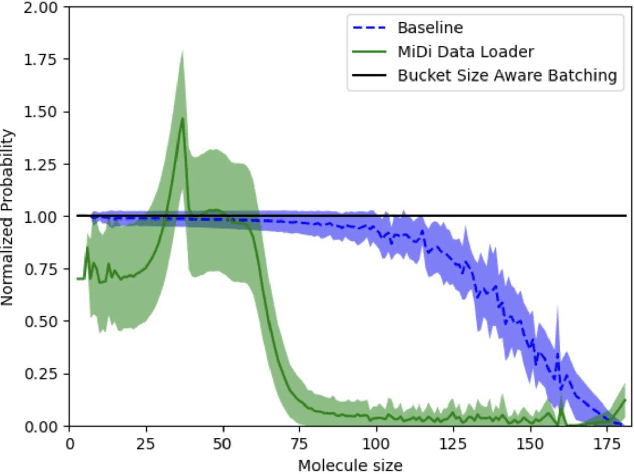
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
EquiJump: Protein Dynamics Simulation via SO(3)-Equivariant Stochastic Interpolants
Oct 12, 2024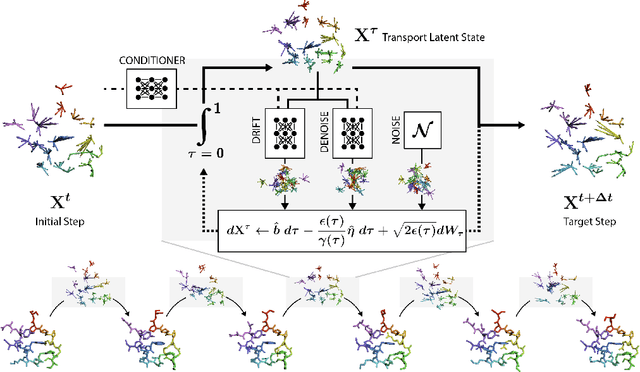
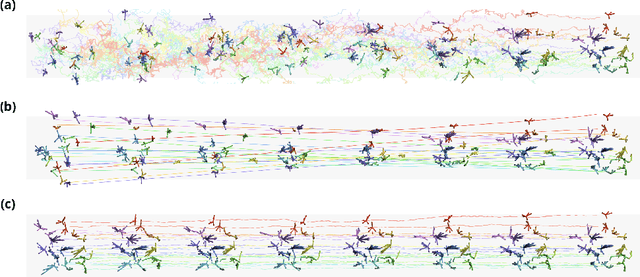
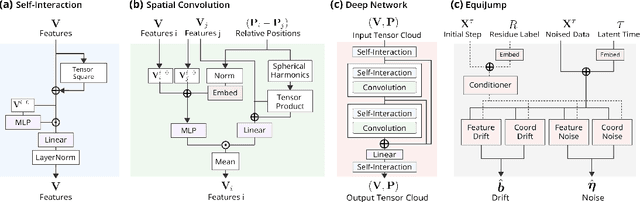
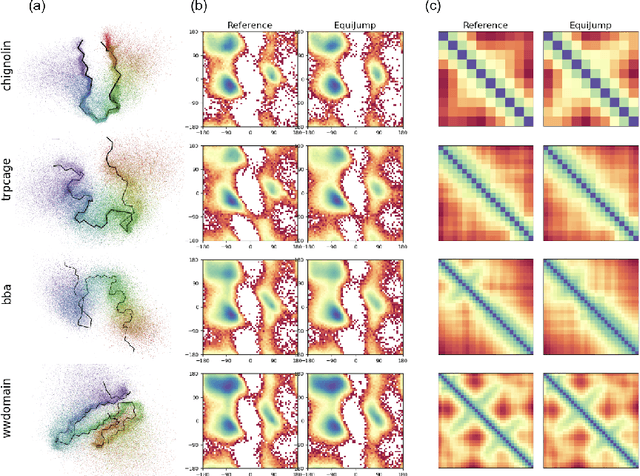
Abstract:Mapping the conformational dynamics of proteins is crucial for elucidating their functional mechanisms. While Molecular Dynamics (MD) simulation enables detailed time evolution of protein motion, its computational toll hinders its use in practice. To address this challenge, multiple deep learning models for reproducing and accelerating MD have been proposed drawing on transport-based generative methods. However, existing work focuses on generation through transport of samples from prior distributions, that can often be distant from the data manifold. The recently proposed framework of stochastic interpolants, instead, enables transport between arbitrary distribution endpoints. Building upon this work, we introduce EquiJump, a transferable SO(3)-equivariant model that bridges all-atom protein dynamics simulation time steps directly. Our approach unifies diverse sampling methods and is benchmarked against existing models on trajectory data of fast folding proteins. EquiJump achieves state-of-the-art results on dynamics simulation with a transferable model on all of the fast folding proteins.
Symphony: Symmetry-Equivariant Point-Centered Spherical Harmonics for Molecule Generation
Nov 27, 2023



Abstract:We present Symphony, an $E(3)$-equivariant autoregressive generative model for 3D molecular geometries that iteratively builds a molecule from molecular fragments. Existing autoregressive models such as G-SchNet and G-SphereNet for molecules utilize rotationally invariant features to respect the 3D symmetries of molecules. In contrast, Symphony uses message-passing with higher-degree $E(3)$-equivariant features. This allows a novel representation of probability distributions via spherical harmonic signals to efficiently model the 3D geometry of molecules. We show that Symphony is able to accurately generate small molecules from the QM9 dataset, outperforming existing autoregressive models and approaching the performance of diffusion models.
Ophiuchus: Scalable Modeling of Protein Structures through Hierarchical Coarse-graining SO(3)-Equivariant Autoencoders
Oct 04, 2023Abstract:Three-dimensional native states of natural proteins display recurring and hierarchical patterns. Yet, traditional graph-based modeling of protein structures is often limited to operate within a single fine-grained resolution, and lacks hourglass neural architectures to learn those high-level building blocks. We narrow this gap by introducing Ophiuchus, an SO(3)-equivariant coarse-graining model that efficiently operates on all heavy atoms of standard protein residues, while respecting their relevant symmetries. Our model departs from current approaches that employ graph modeling, instead focusing on local convolutional coarsening to model sequence-motif interactions in log-linear length complexity. We train Ophiuchus on contiguous fragments of PDB monomers, investigating its reconstruction capabilities across different compression rates. We examine the learned latent space and demonstrate its prompt usage in conformational interpolation, comparing interpolated trajectories to structure snapshots from the PDBFlex dataset. Finally, we leverage denoising diffusion probabilistic models (DDPM) to efficiently sample readily-decodable latent embeddings of diverse miniproteins. Our experiments demonstrate Ophiuchus to be a scalable basis for efficient protein modeling and generation.
A General Framework for Equivariant Neural Networks on Reductive Lie Groups
May 31, 2023Abstract:Reductive Lie Groups, such as the orthogonal groups, the Lorentz group, or the unitary groups, play essential roles across scientific fields as diverse as high energy physics, quantum mechanics, quantum chromodynamics, molecular dynamics, computer vision, and imaging. In this paper, we present a general Equivariant Neural Network architecture capable of respecting the symmetries of the finite-dimensional representations of any reductive Lie Group G. Our approach generalizes the successful ACE and MACE architectures for atomistic point clouds to any data equivariant to a reductive Lie group action. We also introduce the lie-nn software library, which provides all the necessary tools to develop and implement such general G-equivariant neural networks. It implements routines for the reduction of generic tensor products of representations into irreducible representations, making it easy to apply our architecture to a wide range of problems and groups. The generality and performance of our approach are demonstrated by applying it to the tasks of top quark decay tagging (Lorentz group) and shape recognition (orthogonal group).
An end-to-end SE-equivariant segmentation network
Mar 02, 2023Abstract:Convolutional neural networks (CNNs) allow for parameter sharing and translational equivariance by using convolutional kernels in their linear layers. By restricting these kernels to be SO(3)-steerable, CNNs can further improve parameter sharing and equivariance. These equivariant convolutional layers have several advantages over standard convolutional layers, including increased robustness to unseen poses, smaller network size, and improved sample efficiency. Despite this, most segmentation networks used in medical image analysis continue to rely on standard convolutional kernels. In this paper, we present a new family of segmentation networks that use equivariant voxel convolutions based on spherical harmonics, as well as equivariant pooling and normalization operations. These SE(3)-equivariant volumetric segmentation networks, which are robust to data poses not seen during training, do not require rotation-based data augmentation during training. In addition, we demonstrate improved segmentation performance in MRI brain tumor and healthy brain structure segmentation tasks, with enhanced robustness to reduced amounts of training data and improved parameter efficiency. Code to reproduce our results, and to implement the equivariant segmentation networks for other tasks is available at http://github.com/SCAN-NRAD/e3nn_Unet
Dissecting the Effects of SGD Noise in Distinct Regimes of Deep Learning
Jan 31, 2023Abstract:Understanding when the noise in stochastic gradient descent (SGD) affects generalization of deep neural networks remains a challenge, complicated by the fact that networks can operate in distinct training regimes. Here we study how the magnitude of this noise $T$ affects performance as the size of the training set $P$ and the scale of initialization $\alpha$ are varied. For gradient descent, $\alpha$ is a key parameter that controls if the network is `lazy' ($\alpha\gg 1$) or instead learns features ($\alpha\ll 1$). For classification of MNIST and CIFAR10 images, our central results are: (i) obtaining phase diagrams for performance in the $(\alpha,T)$ plane. They show that SGD noise can be detrimental or instead useful depending on the training regime. Moreover, although increasing $T$ or decreasing $\alpha$ both allow the net to escape the lazy regime, these changes can have opposite effects on performance. (ii) Most importantly, we find that key dynamical quantities (including the total variations of weights during training) depend on both $T$ and $P$ as power laws, and the characteristic temperature $T_c$, where the noise of SGD starts affecting performance, is a power law of $P$. These observations indicate that a key effect of SGD noise occurs late in training, by affecting the stopping process whereby all data are fitted. We argue that due to SGD noise, nets must develop a stronger `signal', i.e. larger informative weights, to fit the data, leading to a longer training time. The same effect occurs at larger training set $P$. We confirm this view in the perceptron model, where signal and noise can be precisely measured. Interestingly, exponents characterizing the effect of SGD depend on the density of data near the decision boundary, as we explain.
e3nn: Euclidean Neural Networks
Jul 18, 2022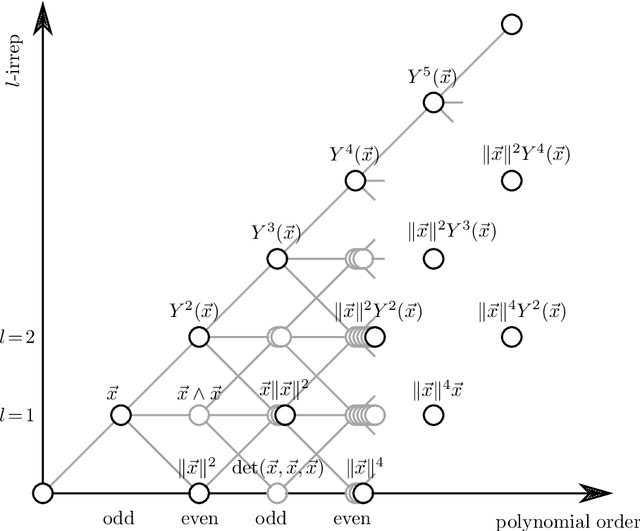
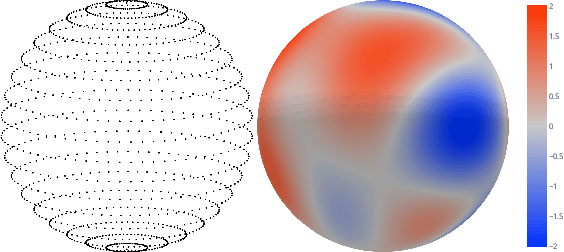
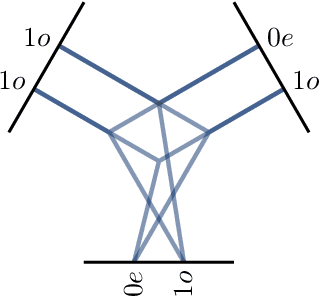
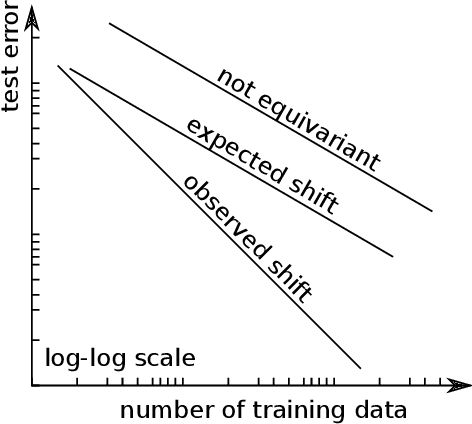
Abstract:We present e3nn, a generalized framework for creating E(3) equivariant trainable functions, also known as Euclidean neural networks. e3nn naturally operates on geometry and geometric tensors that describe systems in 3D and transform predictably under a change of coordinate system. The core of e3nn are equivariant operations such as the TensorProduct class or the spherical harmonics functions that can be composed to create more complex modules such as convolutions and attention mechanisms. These core operations of e3nn can be used to efficiently articulate Tensor Field Networks, 3D Steerable CNNs, Clebsch-Gordan Networks, SE(3) Transformers and other E(3) equivariant networks.
How memory architecture affects performance and learning in simple POMDPs
Jun 16, 2021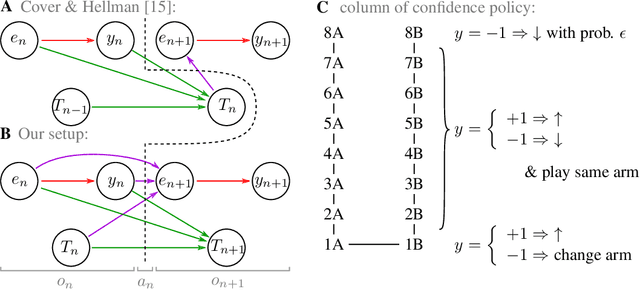
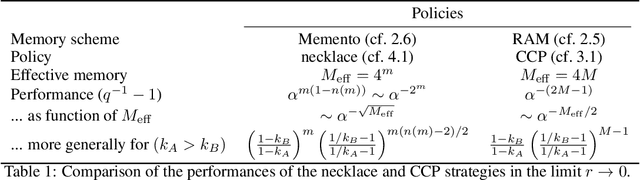
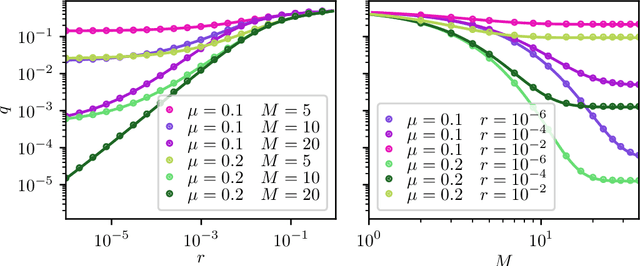
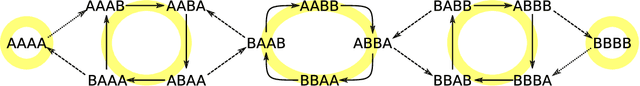
Abstract:Reinforcement learning is made much more complex when the agent's observation is partial or noisy. This case corresponds to a partially observable Markov decision process (POMDP). One strategy to seek good performance in POMDPs is to endow the agent with a finite memory, whose update is governed by the policy. However, policy optimization is non-convex in that case and can lead to poor training performance for random initialization. The performance can be empirically improved by constraining the memory architecture, then sacrificing optimality to facilitate training. Here we study this trade-off in the two-arm bandit problem, and compare two extreme cases: (i) the random access memory where any transitions between $M$ memory states are allowed and (ii) a fixed memory where the agent can access its last $m$ actions and rewards. For (i), the probability $q$ to play the worst arm is known to be exponentially small in $M$ for the optimal policy. Our main result is to show that similar performance can be reached for (ii) as well, despite the simplicity of the memory architecture: using a conjecture on Gray-ordered binary necklaces, we find policies for which $q$ is exponentially small in $2^m$ i.e. $q\sim\alpha^{2^m}$ for some $\alpha < 1$. Interestingly, we observe empirically that training from random initialization leads to very poor results for (i), and significantly better results for (ii).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge