Tess Smidt
PFT: Phonon Fine-tuning for Machine Learned Interatomic Potentials
Jan 12, 2026Abstract:Many materials properties depend on higher-order derivatives of the potential energy surface, yet machine learned interatomic potentials (MLIPs) trained with standard a standard loss on energy, force, and stress errors can exhibit error in curvature, degrading the prediction of vibrational properties. We introduce phonon fine-tuning (PFT), which directly supervises second-order force constants of materials by matching MLIP energy Hessians to DFT-computed force constants from finite displacement phonon calculations. To scale to large supercells, PFT stochastically samples Hessian columns and computes the loss with a single Hessian-vector product. We also use a simple co-training scheme to incorporate upstream data to mitigate catastrophic forgetting. On the MDR Phonon benchmark, PFT improves Nequix MP (trained on Materials Project) by 55% on average across phonon thermodynamic properties and achieves state-of-the-art performance among models trained on Materials Project trajectories. PFT also generalizes to improve properties beyond second-derivatives, improving thermal conductivity predictions that rely on third-order derivatives of the potential energy.
Accelerating Protein Molecular Dynamics Simulation with DeepJump
Sep 16, 2025



Abstract:Unraveling the dynamical motions of biomolecules is essential for bridging their structure and function, yet it remains a major computational challenge. Molecular dynamics (MD) simulation provides a detailed depiction of biomolecular motion, but its high-resolution temporal evolution comes at significant computational cost, limiting its applicability to timescales of biological relevance. Deep learning approaches have emerged as promising solutions to overcome these computational limitations by learning to predict long-timescale dynamics. However, generalizable kinetics models for proteins remain largely unexplored, and the fundamental limits of achievable acceleration while preserving dynamical accuracy are poorly understood. In this work, we fill this gap with DeepJump, an Euclidean-Equivariant Flow Matching-based model for predicting protein conformational dynamics across multiple temporal scales. We train DeepJump on trajectories of the diverse proteins of mdCATH, systematically studying our model's performance in generalizing to long-term dynamics of fast-folding proteins and characterizing the trade-off between computational acceleration and prediction accuracy. We demonstrate the application of DeepJump to ab initio folding, showcasing prediction of folding pathways and native states. Our results demonstrate that DeepJump achieves significant $\approx$1000$\times$ computational acceleration while effectively recovering long-timescale dynamics, providing a stepping stone for enabling routine simulation of proteins.
The Price of Freedom: Exploring Expressivity and Runtime Tradeoffs in Equivariant Tensor Products
Jun 16, 2025Abstract:$E(3)$-equivariant neural networks have demonstrated success across a wide range of 3D modelling tasks. A fundamental operation in these networks is the tensor product, which interacts two geometric features in an equivariant manner to create new features. Due to the high computational complexity of the tensor product, significant effort has been invested to optimize the runtime of this operation. For example, Luo et al. (2024) recently proposed the Gaunt tensor product (GTP) which promises a significant speedup. In this work, we provide a careful, systematic analysis of a number of tensor product operations. In particular, we emphasize that different tensor products are not performing the same operation. The reported speedups typically come at the cost of expressivity. We introduce measures of expressivity and interactability to characterize these differences. In addition, we realized the original implementation of GTP can be greatly simplified by directly using a spherical grid at no cost in asymptotic runtime. This spherical grid approach is faster on our benchmarks and in actual training of the MACE interatomic potential by 30\%. Finally, we provide the first systematic microbenchmarks of the various tensor product operations. We find that the theoretical runtime guarantees can differ wildly from empirical performance, demonstrating the need for careful application-specific benchmarking. Code is available at \href{https://github.com/atomicarchitects/PriceofFreedom}{https://github.com/atomicarchitects/PriceofFreedom}
High-performance training and inference for deep equivariant interatomic potentials
Apr 22, 2025Abstract:Machine learning interatomic potentials, particularly those based on deep equivariant neural networks, have demonstrated state-of-the-art accuracy and computational efficiency in atomistic modeling tasks like molecular dynamics and high-throughput screening. The size of datasets and demands of downstream workflows are growing rapidly, making robust and scalable software essential. This work presents a major overhaul of the NequIP framework focusing on multi-node parallelism, computational performance, and extensibility. The redesigned framework supports distributed training on large datasets and removes barriers preventing full utilization of the PyTorch 2.0 compiler at train time. We demonstrate this acceleration in a case study by training Allegro models on the SPICE 2 dataset of organic molecular systems. For inference, we introduce the first end-to-end infrastructure that uses the PyTorch Ahead-of-Time Inductor compiler for machine learning interatomic potentials. Additionally, we implement a custom kernel for the Allegro model's most expensive operation, the tensor product. Together, these advancements speed up molecular dynamics calculations on system sizes of practical relevance by up to a factor of 18.
A Cosmic-Scale Benchmark for Symmetry-Preserving Data Processing
Oct 27, 2024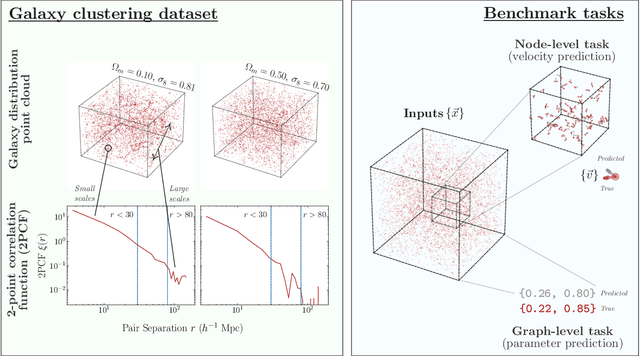
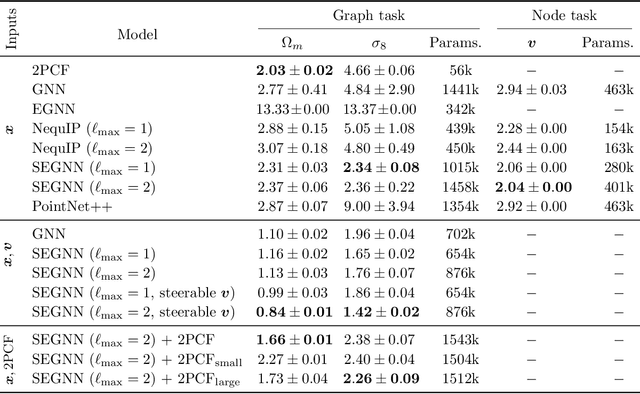
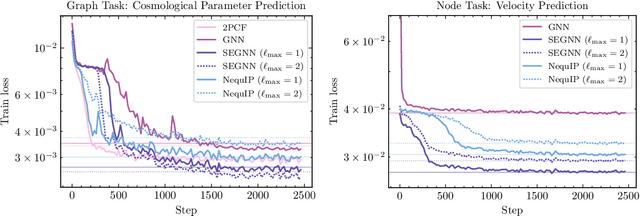

Abstract:Efficiently processing structured point cloud data while preserving multiscale information is a key challenge across domains, from graphics to atomistic modeling. Using a curated dataset of simulated galaxy positions and properties, represented as point clouds, we benchmark the ability of graph neural networks to simultaneously capture local clustering environments and long-range correlations. Given the homogeneous and isotropic nature of the Universe, the data exhibits a high degree of symmetry. We therefore focus on evaluating the performance of Euclidean symmetry-preserving ($E(3)$-equivariant) graph neural networks, showing that they can outperform non-equivariant counterparts and domain-specific information extraction techniques in downstream performance as well as simulation-efficiency. However, we find that current architectures fail to capture information from long-range correlations as effectively as domain-specific baselines, motivating future work on architectures better suited for extracting long-range information.
EquiJump: Protein Dynamics Simulation via SO(3)-Equivariant Stochastic Interpolants
Oct 12, 2024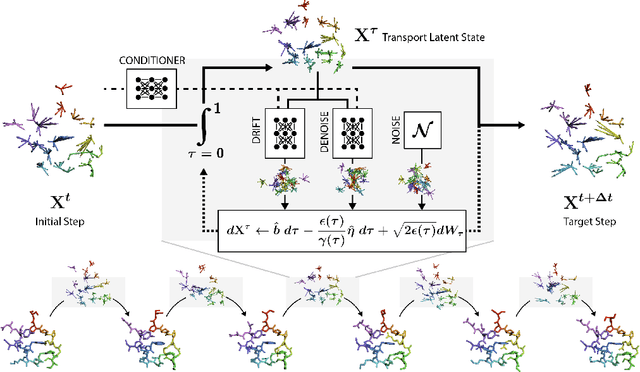
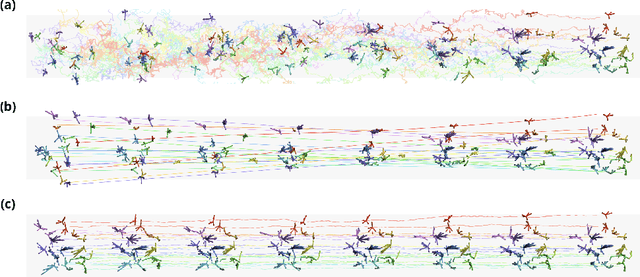
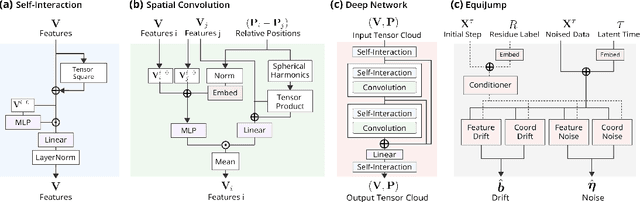
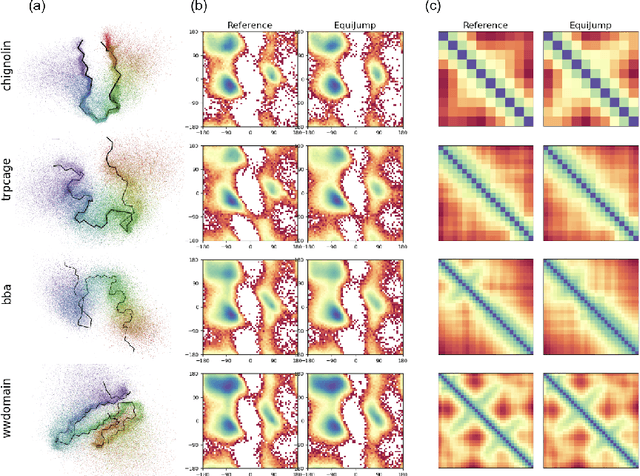
Abstract:Mapping the conformational dynamics of proteins is crucial for elucidating their functional mechanisms. While Molecular Dynamics (MD) simulation enables detailed time evolution of protein motion, its computational toll hinders its use in practice. To address this challenge, multiple deep learning models for reproducing and accelerating MD have been proposed drawing on transport-based generative methods. However, existing work focuses on generation through transport of samples from prior distributions, that can often be distant from the data manifold. The recently proposed framework of stochastic interpolants, instead, enables transport between arbitrary distribution endpoints. Building upon this work, we introduce EquiJump, a transferable SO(3)-equivariant model that bridges all-atom protein dynamics simulation time steps directly. Our approach unifies diverse sampling methods and is benchmarked against existing models on trajectory data of fast folding proteins. EquiJump achieves state-of-the-art results on dynamics simulation with a transferable model on all of the fast folding proteins.
Relaxed Equivariant Graph Neural Networks
Jul 30, 2024Abstract:3D Euclidean symmetry equivariant neural networks have demonstrated notable success in modeling complex physical systems. We introduce a framework for relaxed $E(3)$ graph equivariant neural networks that can learn and represent symmetry breaking within continuous groups. Building on the existing e3nn framework, we propose the use of relaxed weights to allow for controlled symmetry breaking. We show empirically that these relaxed weights learn the correct amount of symmetry breaking.
A Recipe for Charge Density Prediction
May 29, 2024Abstract:In density functional theory, charge density is the core attribute of atomic systems from which all chemical properties can be derived. Machine learning methods are promising in significantly accelerating charge density prediction, yet existing approaches either lack accuracy or scalability. We propose a recipe that can achieve both. In particular, we identify three key ingredients: (1) representing the charge density with atomic and virtual orbitals (spherical fields centered at atom/virtual coordinates); (2) using expressive and learnable orbital basis sets (basis function for the spherical fields); and (3) using high-capacity equivariant neural network architecture. Our method achieves state-of-the-art accuracy while being more than an order of magnitude faster than existing methods. Furthermore, our method enables flexible efficiency-accuracy trade-offs by adjusting the model/basis sizes.
Generalizing Denoising to Non-Equilibrium Structures Improves Equivariant Force Fields
Mar 14, 2024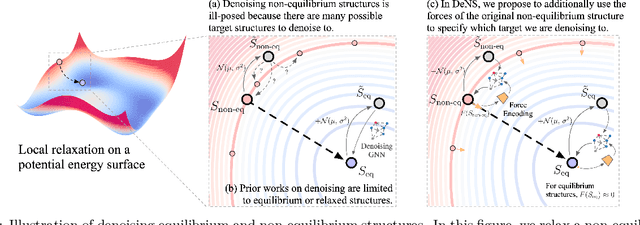
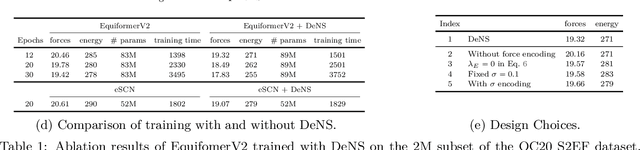
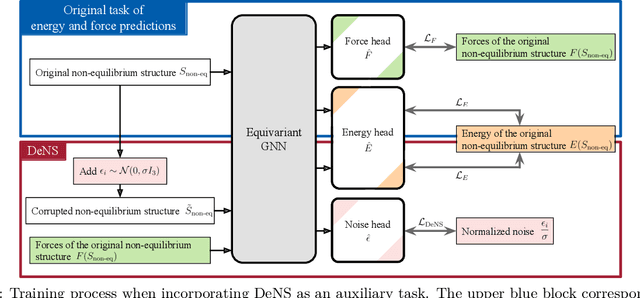
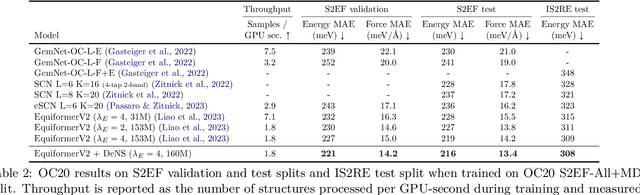
Abstract:Understanding the interactions of atoms such as forces in 3D atomistic systems is fundamental to many applications like molecular dynamics and catalyst design. However, simulating these interactions requires compute-intensive ab initio calculations and thus results in limited data for training neural networks. In this paper, we propose to use denoising non-equilibrium structures (DeNS) as an auxiliary task to better leverage training data and improve performance. For training with DeNS, we first corrupt a 3D structure by adding noise to its 3D coordinates and then predict the noise. Different from previous works on denoising, which are limited to equilibrium structures, the proposed method generalizes denoising to a much larger set of non-equilibrium structures. The main difference is that a non-equilibrium structure does not correspond to local energy minima and has non-zero forces, and therefore it can have many possible atomic positions compared to an equilibrium structure. This makes denoising non-equilibrium structures an ill-posed problem since the target of denoising is not uniquely defined. Our key insight is to additionally encode the forces of the original non-equilibrium structure to specify which non-equilibrium structure we are denoising. Concretely, given a corrupted non-equilibrium structure and the forces of the original one, we predict the non-equilibrium structure satisfying the input forces instead of any arbitrary structures. Since DeNS requires encoding forces, DeNS favors equivariant networks, which can easily incorporate forces and other higher-order tensors in node embeddings. We study the effectiveness of training equivariant networks with DeNS on OC20, OC22 and MD17 datasets and demonstrate that DeNS can achieve new state-of-the-art results on OC20 and OC22 and significantly improve training efficiency on MD17.
Equivariant Symmetry Breaking Sets
Feb 05, 2024



Abstract:Equivariant neural networks (ENNs) have been shown to be extremely effective in applications involving underlying symmetries. By construction ENNs cannot produce lower symmetry outputs given a higher symmetry input. However, spontaneous symmetry breaking occurs in many physical systems and we may obtain a less symmetric stable state from an initial highly symmetric one. Hence, it is imperative that we understand how to systematically break symmetry in ENNs. In this work, we propose a novel symmetry breaking framework that is fully equivariant. We emphasize that our approach is general and applicable to equivariance under any group. To achieve this, we introduce the idea of symmetry breaking sets (SBS). Rather than redesign existing networks, we design sets of symmetry breaking objects which we feed into our network based on the symmetry of our inputs and outputs. We show there is a natural way to define equivariance on these sets, which gives an additional constraint. Minimizing the size of these sets equates to data efficiency. We prove that minimizing these sets translates to a well studied group theory problem, and tabulate solutions to this problem for the point groups. Finally, we provide some examples of symmetry breaking to demonstrate how our approach works in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge