Lukas Picek
GeoPl@ntNet: A Platform for Exploring Essential Biodiversity Variables
Nov 16, 2025Abstract:This paper describes GeoPl@ntNet, an interactive web application designed to make Essential Biodiversity Variables accessible and understandable to everyone through dynamic maps and fact sheets. Its core purpose is to allow users to explore high-resolution AI-generated maps of species distributions, habitat types, and biodiversity indicators across Europe. These maps, developed through a cascading pipeline involving convolutional neural networks and large language models, provide an intuitive yet information-rich interface to better understand biodiversity, with resolutions as precise as 50x50 meters. The website also enables exploration of specific regions, allowing users to select areas of interest on the map (e.g., urban green spaces, protected areas, or riverbanks) to view local species and their coverage. Additionally, GeoPl@ntNet generates comprehensive reports for selected regions, including insights into the number of protected species, invasive species, and endemic species.
CzechLynx: A Dataset for Individual Identification and Pose Estimation of the Eurasian Lynx
Jun 05, 2025Abstract:We introduce CzechLynx, the first large-scale, open-access dataset for individual identification, 2D pose estimation, and instance segmentation of the Eurasian lynx (Lynx lynx). CzechLynx includes more than 30k camera trap images annotated with segmentation masks, identity labels, and 20-point skeletons and covers 219 unique individuals across 15 years of systematic monitoring in two geographically distinct regions: Southwest Bohemia and the Western Carpathians. To increase the data variability, we create a complementary synthetic set with more than 100k photorealistic images generated via a Unity-based pipeline and diffusion-driven text-to-texture modeling, covering diverse environments, poses, and coat-pattern variations. To allow testing generalization across spatial and temporal domains, we define three tailored evaluation protocols/splits: (i) geo-aware, (ii) time-aware open-set, and (iii) time-aware closed-set. This dataset is targeted to be instrumental in benchmarking state-of-the-art models and the development of novel methods for not just individual animal re-identification.
Mapping biodiversity at very-high resolution in Europe
Apr 07, 2025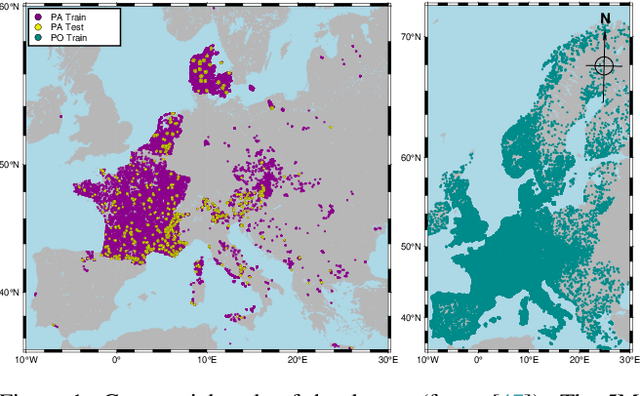
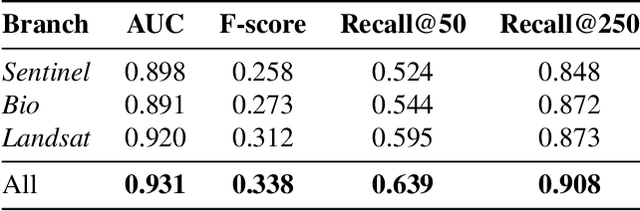
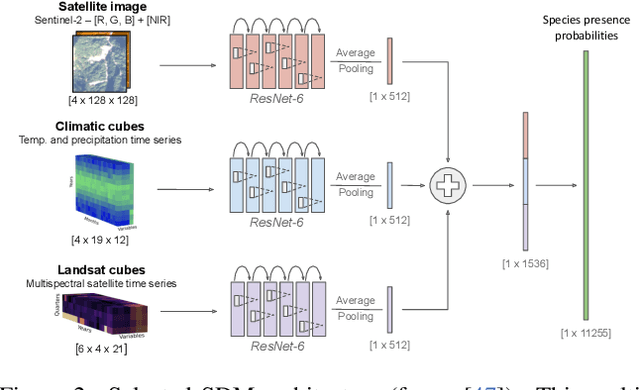

Abstract:This paper describes a cascading multimodal pipeline for high-resolution biodiversity mapping across Europe, integrating species distribution modeling, biodiversity indicators, and habitat classification. The proposed pipeline first predicts species compositions using a deep-SDM, a multimodal model trained on remote sensing, climate time series, and species occurrence data at 50x50m resolution. These predictions are then used to generate biodiversity indicator maps and classify habitats with Pl@ntBERT, a transformer-based LLM designed for species-to-habitat mapping. With this approach, continental-scale species distribution maps, biodiversity indicator maps, and habitat maps are produced, providing fine-grained ecological insights. Unlike traditional methods, this framework enables joint modeling of interspecies dependencies, bias-aware training with heterogeneous presence-absence data, and large-scale inference from multi-source remote sensing inputs.
Zero-shot Hazard Identification in Autonomous Driving: A Case Study on the COOOL Benchmark
Dec 27, 2024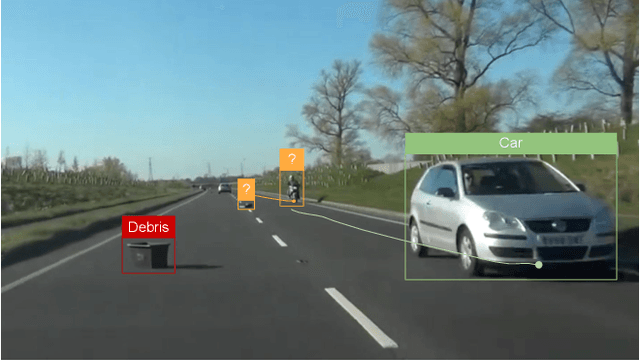
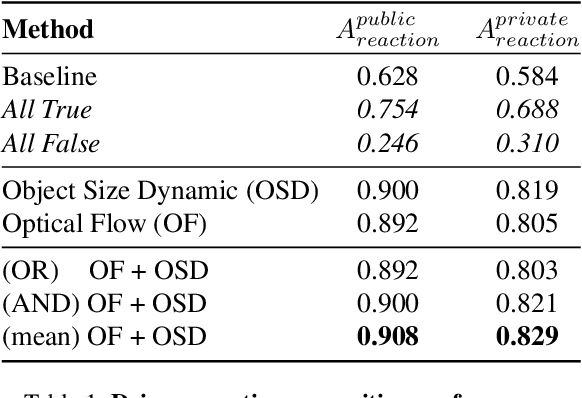
Abstract:This paper presents our submission to the COOOL competition, a novel benchmark for detecting and classifying out-of-label hazards in autonomous driving. Our approach integrates diverse methods across three core tasks: (i) driver reaction detection, (ii) hazard object identification, and (iii) hazard captioning. We propose kernel-based change point detection on bounding boxes and optical flow dynamics for driver reaction detection to analyze motion patterns. For hazard identification, we combined a naive proximity-based strategy with object classification using a pre-trained ViT model. At last, for hazard captioning, we used the MOLMO vision-language model with tailored prompts to generate precise and context-aware descriptions of rare and low-resolution hazards. The proposed pipeline outperformed the baseline methods by a large margin, reducing the relative error by 33%, and scored 2nd on the final leaderboard consisting of 32 teams.
Towards Zero-Shot Camera Trap Image Categorization
Oct 16, 2024



Abstract:This paper describes the search for an alternative approach to the automatic categorization of camera trap images. First, we benchmark state-of-the-art classifiers using a single model for all images. Next, we evaluate methods combining MegaDetector with one or more classifiers and Segment Anything to assess their impact on reducing location-specific overfitting. Last, we propose and test two approaches using large language and foundational models, such as DINOv2, BioCLIP, BLIP, and ChatGPT, in a zero-shot scenario. Evaluation carried out on two publicly available datasets (WCT from New Zealand, CCT20 from the Southwestern US) and a private dataset (CEF from Central Europe) revealed that combining MegaDetector with two separate classifiers achieves the highest accuracy. This approach reduced the relative error of a single BEiTV2 classifier by approximately 42\% on CCT20, 48\% on CEF, and 75\% on WCT. Besides, as the background is removed, the error in terms of accuracy in new locations is reduced to half. The proposed zero-shot pipeline based on DINOv2 and FAISS achieved competitive results (1.0\% and 4.7\% smaller on CCT20, and CEF, respectively), which highlights the potential of zero-shot approaches for camera trap image categorization.
MALPOLON: A Framework for Deep Species Distribution Modeling
Sep 26, 2024Abstract:This paper describes a deep-SDM framework, MALPOLON. Written in Python and built upon the PyTorch library, this framework aims to facilitate training and inferences of deep species distribution models (deep-SDM) and sharing for users with only general Python language skills (e.g., modeling ecologists) who are interested in testing deep learning approaches to build new SDMs. More advanced users can also benefit from the framework's modularity to run more specific experiments by overriding existing classes while taking advantage of press-button examples to train neural networks on multiple classification tasks using custom or provided raw and pre-processed datasets. The framework is open-sourced on GitHub and PyPi along with extensive documentation and examples of use in various scenarios. MALPOLON offers straightforward installation, YAML-based configuration, parallel computing, multi-GPU utilization, baseline and foundational models for benchmarking, and extensive tutorials/documentation, aiming to enhance accessibility and performance scalability for ecologists and researchers.
GeoPlant: Spatial Plant Species Prediction Dataset
Aug 25, 2024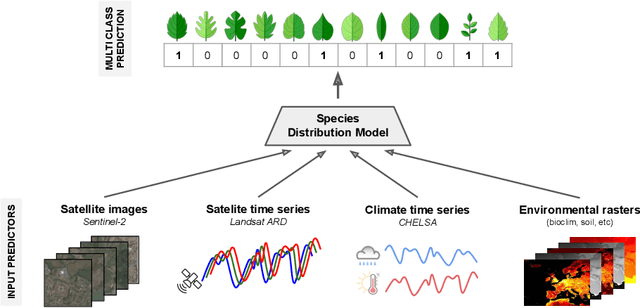
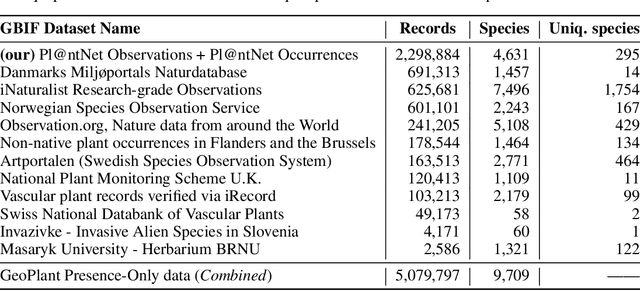
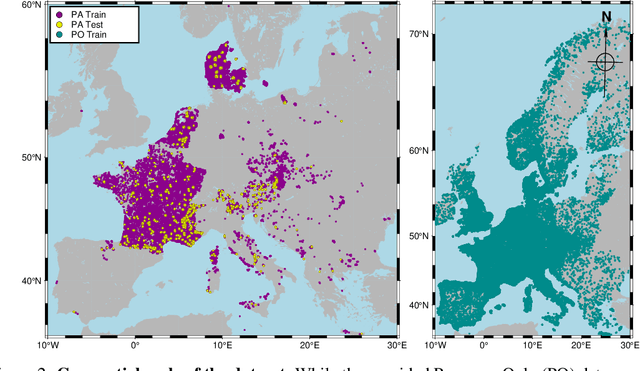
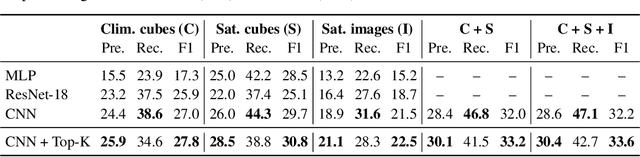
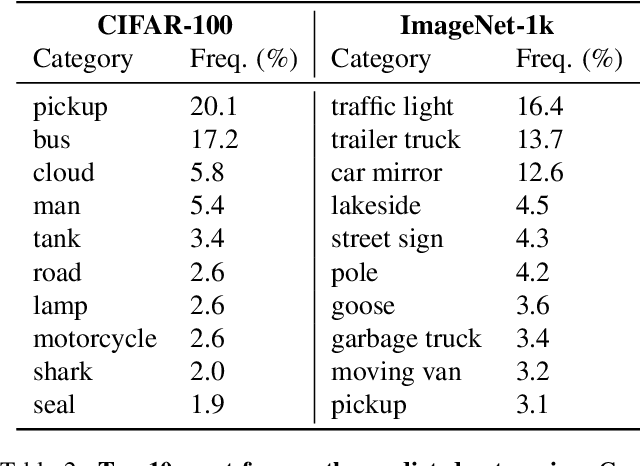
Abstract:The difficulty of monitoring biodiversity at fine scales and over large areas limits ecological knowledge and conservation efforts. To fill this gap, Species Distribution Models (SDMs) predict species across space from spatially explicit features. Yet, they face the challenge of integrating the rich but heterogeneous data made available over the past decade, notably millions of opportunistic species observations and standardized surveys, as well as multi-modal remote sensing data. In light of that, we have designed and developed a new European-scale dataset for SDMs at high spatial resolution (10-50 m), including more than 10k species (i.e., most of the European flora). The dataset comprises 5M heterogeneous Presence-Only records and 90k exhaustive Presence-Absence survey records, all accompanied by diverse environmental rasters (e.g., elevation, human footprint, and soil) that are traditionally used in SDMs. In addition, it provides Sentinel-2 RGB and NIR satellite images with 10 m resolution, a 20-year time-series of climatic variables, and satellite time-series from the Landsat program. In addition to the data, we provide an openly accessible SDM benchmark (hosted on Kaggle), which has already attracted an active community and a set of strong baselines for single predictor/modality and multimodal approaches. All resources, e.g., the dataset, pre-trained models, and baseline methods (in the form of notebooks), are available on Kaggle, allowing one to start with our dataset literally with two mouse clicks.
FungiTastic: A multi-modal dataset and benchmark for image categorization
Aug 24, 2024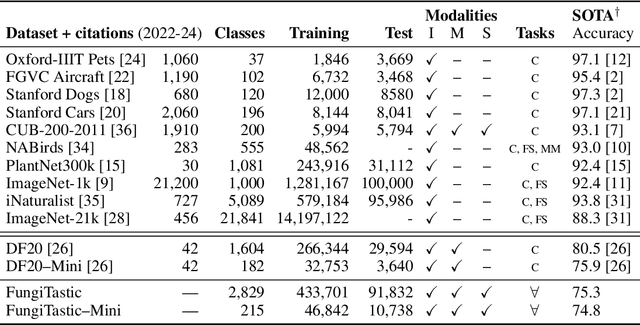
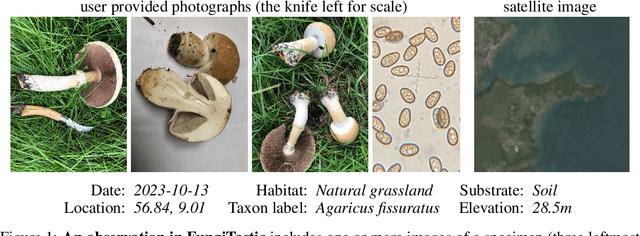
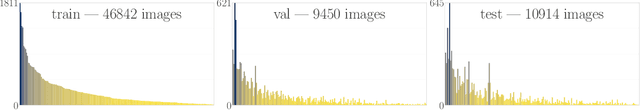
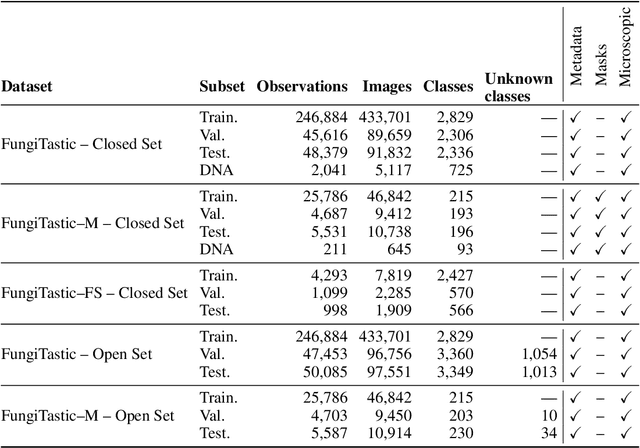
Abstract:We introduce a new, highly challenging benchmark and a dataset -- FungiTastic -- based on data continuously collected over a twenty-year span. The dataset originates in fungal records labeled and curated by experts. It consists of about 350k multi-modal observations that include more than 650k photographs from 5k fine-grained categories and diverse accompanying information, e.g., acquisition metadata, satellite images, and body part segmentation. FungiTastic is the only benchmark that includes a test set with partially DNA-sequenced ground truth of unprecedented label reliability. The benchmark is designed to support (i) standard close-set classification, (ii) open-set classification, (iii) multi-modal classification, (iv) few-shot learning, (v) domain shift, and many more. We provide baseline methods tailored for almost all the use-cases. We provide a multitude of ready-to-use pre-trained models on HuggingFace and a framework for model training. A comprehensive documentation describing the dataset features and the baselines are available at https://bohemianvra.github.io/FungiTastic/ and https://www.kaggle.com/datasets/picekl/fungitastic.
WildFusion: Individual Animal Identification with Calibrated Similarity Fusion
Aug 23, 2024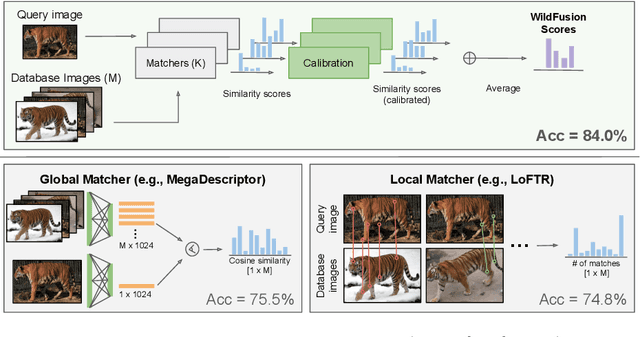
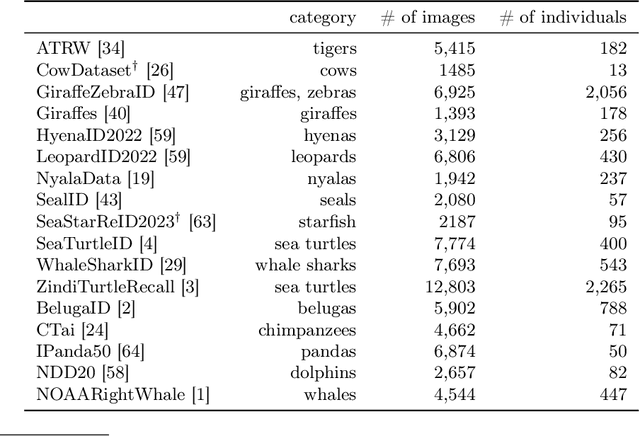

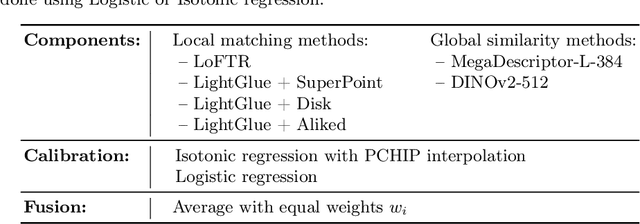
Abstract:We propose a new method - WildFusion - for individual identification of a broad range of animal species. The method fuses deep scores (e.g., MegaDescriptor or DINOv2) and local matching similarity (e.g., LoFTR and LightGlue) to identify individual animals. The global and local information fusion is facilitated by similarity score calibration. In a zero-shot setting, relying on local similarity score only, WildFusion achieved mean accuracy, measured on 17 datasets, of 76.2%. This is better than the state-of-the-art model, MegaDescriptor-L, whose training set included 15 of the 17 datasets. If a dataset-specific calibration is applied, mean accuracy increases by 2.3% percentage points. WildFusion, with both local and global similarity scores, outperforms the state-of-the-art significantly - mean accuracy reached 84.0%, an increase of 8.5 percentage points; the mean relative error drops by 35%. We make the code and pre-trained models publicly available5, enabling immediate use in ecology and conservation.
Animal Identification with Independent Foreground and Background Modeling
Aug 23, 2024



Abstract:We propose a method that robustly exploits background and foreground in visual identification of individual animals. Experiments show that their automatic separation, made easy with methods like Segment Anything, together with independent foreground and background-related modeling, improves results. The two predictions are combined in a principled way, thanks to novel Per-Instance Temperature Scaling that helps the classifier to deal with appearance ambiguities in training and to produce calibrated outputs in the inference phase. For identity prediction from the background, we propose novel spatial and temporal models. On two problems, the relative error w.r.t. the baseline was reduced by 22.3% and 8.8%, respectively. For cases where objects appear in new locations, an example of background drift, accuracy doubles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge