Benjamin Deneu
LIRMM, ZENITH
Mapping biodiversity at very-high resolution in Europe
Apr 07, 2025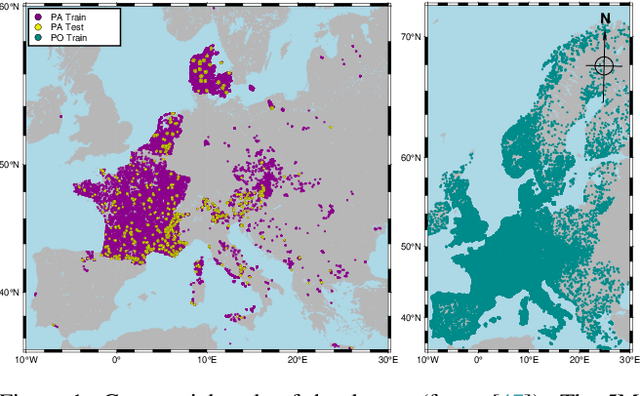

Abstract:This paper describes a cascading multimodal pipeline for high-resolution biodiversity mapping across Europe, integrating species distribution modeling, biodiversity indicators, and habitat classification. The proposed pipeline first predicts species compositions using a deep-SDM, a multimodal model trained on remote sensing, climate time series, and species occurrence data at 50x50m resolution. These predictions are then used to generate biodiversity indicator maps and classify habitats with Pl@ntBERT, a transformer-based LLM designed for species-to-habitat mapping. With this approach, continental-scale species distribution maps, biodiversity indicator maps, and habitat maps are produced, providing fine-grained ecological insights. Unlike traditional methods, this framework enables joint modeling of interspecies dependencies, bias-aware training with heterogeneous presence-absence data, and large-scale inference from multi-source remote sensing inputs.
MALPOLON: A Framework for Deep Species Distribution Modeling
Sep 26, 2024Abstract:This paper describes a deep-SDM framework, MALPOLON. Written in Python and built upon the PyTorch library, this framework aims to facilitate training and inferences of deep species distribution models (deep-SDM) and sharing for users with only general Python language skills (e.g., modeling ecologists) who are interested in testing deep learning approaches to build new SDMs. More advanced users can also benefit from the framework's modularity to run more specific experiments by overriding existing classes while taking advantage of press-button examples to train neural networks on multiple classification tasks using custom or provided raw and pre-processed datasets. The framework is open-sourced on GitHub and PyPi along with extensive documentation and examples of use in various scenarios. MALPOLON offers straightforward installation, YAML-based configuration, parallel computing, multi-GPU utilization, baseline and foundational models for benchmarking, and extensive tutorials/documentation, aiming to enhance accessibility and performance scalability for ecologists and researchers.
GeoPlant: Spatial Plant Species Prediction Dataset
Aug 25, 2024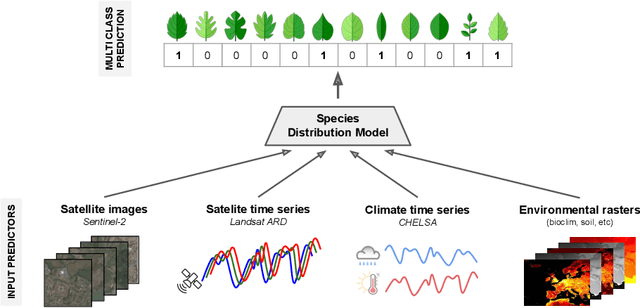
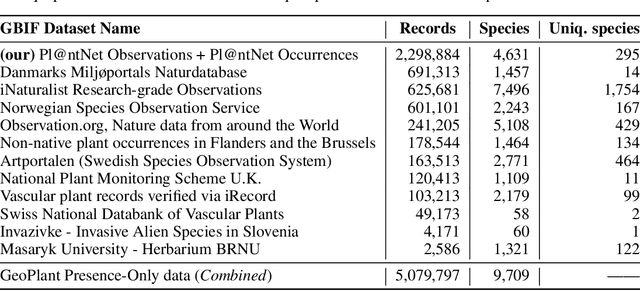
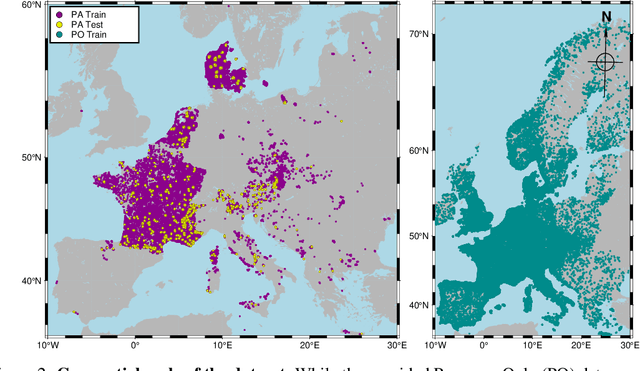
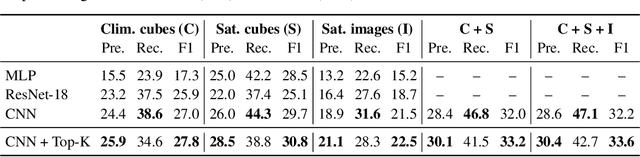
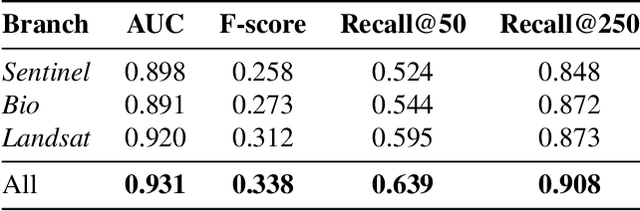
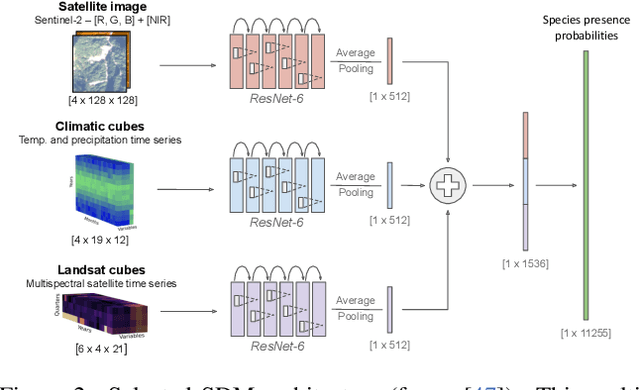
Abstract:The difficulty of monitoring biodiversity at fine scales and over large areas limits ecological knowledge and conservation efforts. To fill this gap, Species Distribution Models (SDMs) predict species across space from spatially explicit features. Yet, they face the challenge of integrating the rich but heterogeneous data made available over the past decade, notably millions of opportunistic species observations and standardized surveys, as well as multi-modal remote sensing data. In light of that, we have designed and developed a new European-scale dataset for SDMs at high spatial resolution (10-50 m), including more than 10k species (i.e., most of the European flora). The dataset comprises 5M heterogeneous Presence-Only records and 90k exhaustive Presence-Absence survey records, all accompanied by diverse environmental rasters (e.g., elevation, human footprint, and soil) that are traditionally used in SDMs. In addition, it provides Sentinel-2 RGB and NIR satellite images with 10 m resolution, a 20-year time-series of climatic variables, and satellite time-series from the Landsat program. In addition to the data, we provide an openly accessible SDM benchmark (hosted on Kaggle), which has already attracted an active community and a set of strong baselines for single predictor/modality and multimodal approaches. All resources, e.g., the dataset, pre-trained models, and baseline methods (in the form of notebooks), are available on Kaggle, allowing one to start with our dataset literally with two mouse clicks.
The GeoLifeCLEF 2020 Dataset
Apr 08, 2020
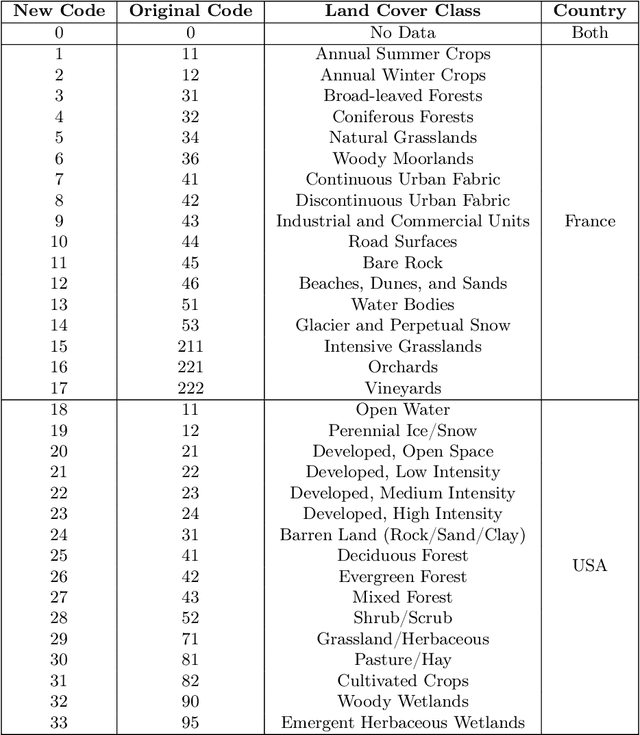
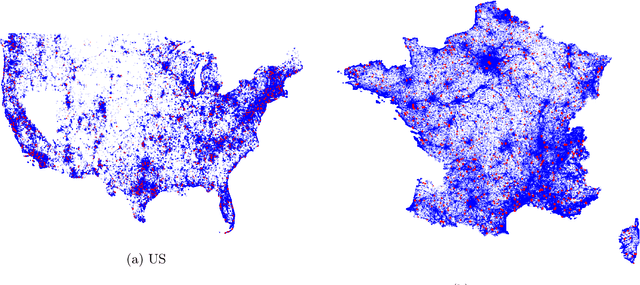
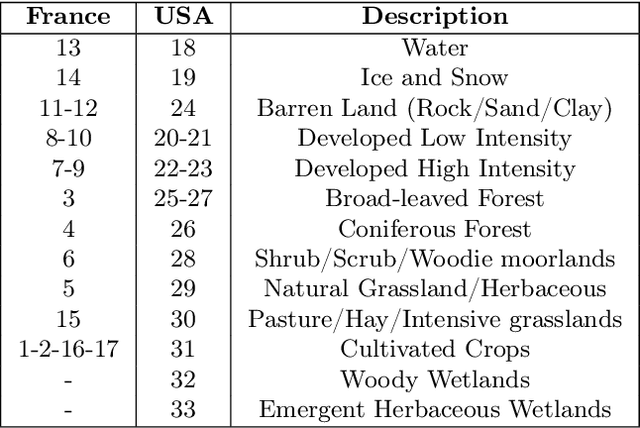
Abstract:Understanding the geographic distribution of species is a key concern in conservation. By pairing species occurrences with environmental features, researchers can model the relationship between an environment and the species which may be found there. To facilitate research in this area, we present the GeoLifeCLEF 2020 dataset, which consists of 1.9 million species observations paired with high-resolution remote sensing imagery, land cover data, and altitude, in addition to traditional low-resolution climate and soil variables. We also discuss the GeoLifeCLEF 2020 competition, which aims to use this dataset to advance the state-of-the-art in location-based species recommendation.
Evaluation of Deep Species Distribution Models using Environment and Co-occurrences
Sep 19, 2019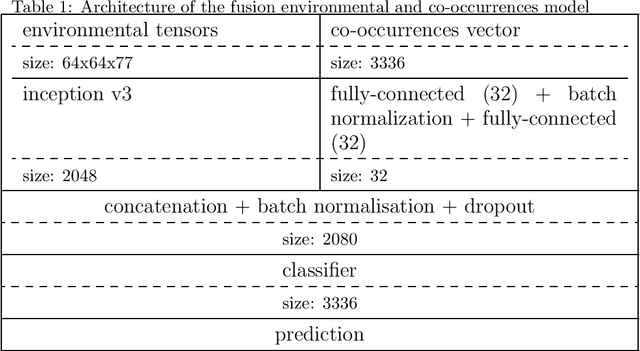
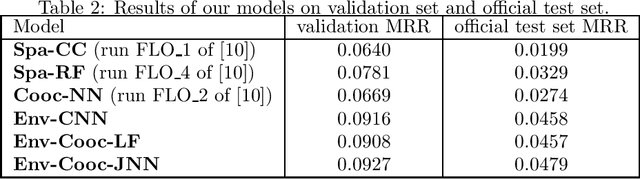
Abstract:This paper presents an evaluation of several approaches of plants species distribution modeling based on spatial, environmental and co-occurrences data using machine learning methods. In particular, we re-evaluate the environmental convolutional neural network model that obtained the best performance of the GeoLifeCLEF 2018 challenge but on a revised dataset that fixes some of the issues of the previous one. We also go deeper in the analysis of co-occurrences information by evaluating a new model that jointly takes environmental variables and co-occurrences as inputs of an end-to-end network. Results show that the environmental models are the best performing methods and that there is a significant amount of complementary information between co-occurrences and environment. Indeed, the model learned on both inputs allows a significant performance gain compared to the environmental model alone.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge