Kostas Papafitsoros
Learning Spatially Adaptive $\ell_1$-Norms Weights for Convolutional Synthesis Regularization
Mar 12, 2025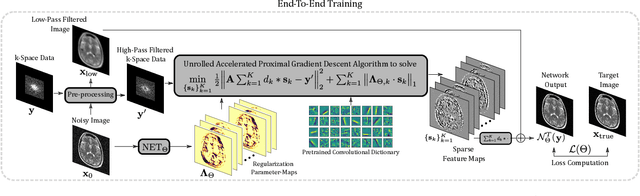
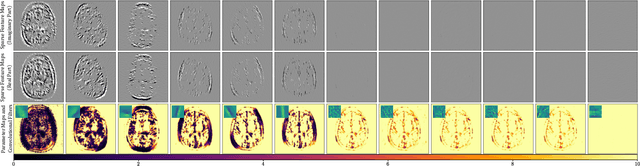
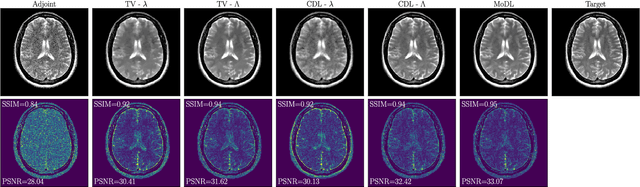
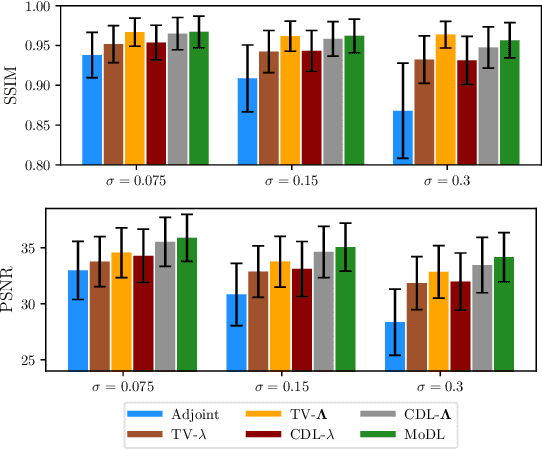
Abstract:We propose an unrolled algorithm approach for learning spatially adaptive parameter maps in the framework of convolutional synthesis-based $\ell_1$ regularization. More precisely, we consider a family of pre-trained convolutional filters and estimate deeply parametrized spatially varying parameters applied to the sparse feature maps by means of unrolling a FISTA algorithm to solve the underlying sparse estimation problem. The proposed approach is evaluated for image reconstruction of low-field MRI and compared to spatially adaptive and non-adaptive analysis-type procedures relying on Total Variation regularization and to a well-established model-based deep learning approach. We show that the proposed approach produces visually and quantitatively comparable results with the latter approaches and at the same time remains highly interpretable. In particular, the inferred parameter maps quantify the local contribution of each filter in the reconstruction, which provides valuable insight into the algorithm mechanism and could potentially be used to discard unsuited filters.
Deep unrolling for learning optimal spatially varying regularisation parameters for Total Generalised Variation
Feb 23, 2025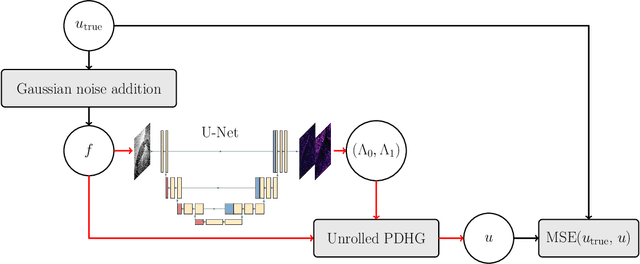
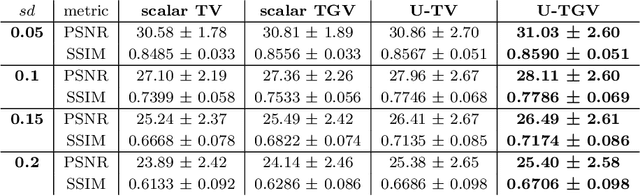
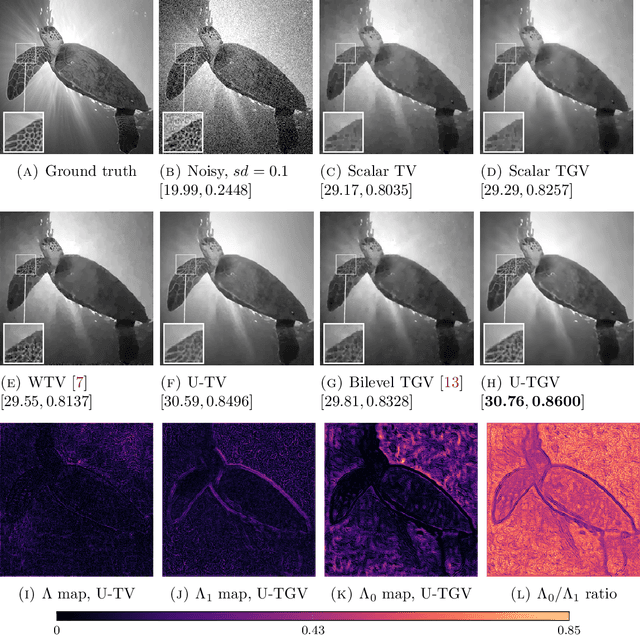
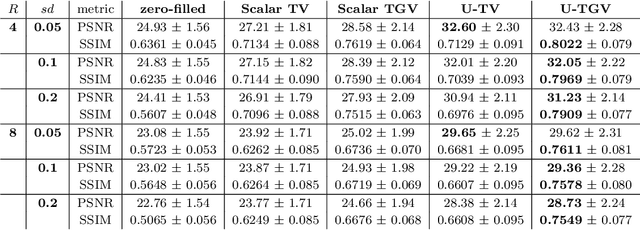
Abstract:We extend a recently introduced deep unrolling framework for learning spatially varying regularisation parameters in inverse imaging problems to the case of Total Generalised Variation (TGV). The framework combines a deep convolutional neural network (CNN) inferring the two spatially varying TGV parameters with an unrolled algorithmic scheme that solves the corresponding variational problem. The two subnetworks are jointly trained end-to-end in a supervised fashion and as such the CNN learns to compute those parameters that drive the reconstructed images as close to the ground truth as possible. Numerical results in image denoising and MRI reconstruction show a significant qualitative and quantitative improvement compared to the best TGV scalar parameter case as well as to other approaches employing spatially varying parameters computed by unsupervised methods. We also observe that the inferred spatially varying parameter maps have a consistent structure near the image edges, asking for further theoretical investigations. In particular, the parameter that weighs the first-order TGV term has a triple-edge structure with alternating high-low-high values whereas the one that weighs the second-order term attains small values in a large neighbourhood around the edges.
Why do we regularise in every iteration for imaging inverse problems?
Nov 01, 2024

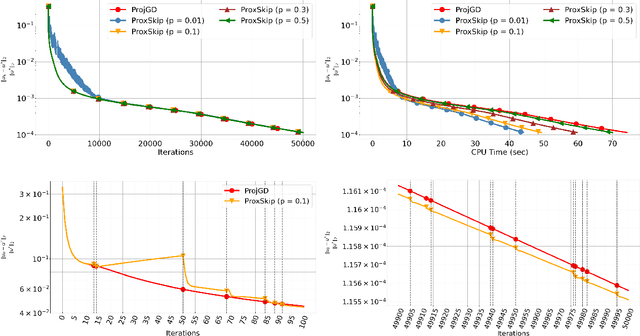
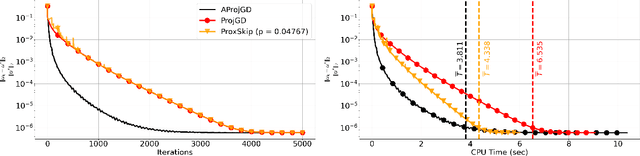
Abstract:Regularisation is commonly used in iterative methods for solving imaging inverse problems. Many algorithms involve the evaluation of the proximal operator of the regularisation term in every iteration, leading to a significant computational overhead since such evaluation can be costly. In this context, the ProxSkip algorithm, recently proposed for federated learning purposes, emerges as an solution. It randomly skips regularisation steps, reducing the computational time of an iterative algorithm without affecting its convergence. Here we explore for the first time the efficacy of ProxSkip to a variety of imaging inverse problems and we also propose a novel PDHGSkip version. Extensive numerical results highlight the potential of these methods to accelerate computations while maintaining high-quality reconstructions.
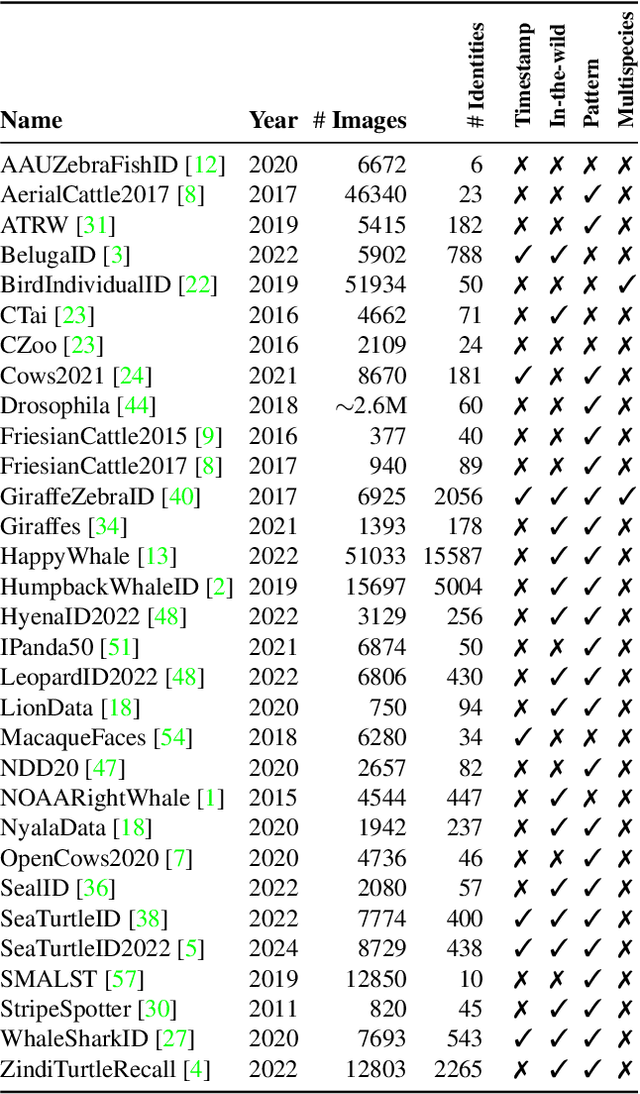
WildlifeReID-10k: Wildlife re-identification dataset with 10k individual animals
Jun 17, 2024Abstract:We introduce a new wildlife re-identification dataset WildlifeReID-10k with more than 214k images of 10k individual animals. It is a collection of 30 existing wildlife re-identification datasets with additional processing steps. WildlifeReID-10k contains animals as diverse as marine turtles, primates, birds, African herbivores, marine mammals and domestic animals. Due to the ubiquity of similar images in datasets, we argue that the standard (random) splits into training and testing sets are inadequate for wildlife re-identification and propose a new similarity-aware split based on the similarity of extracted features. To promote fair method comparison, we include similarity-aware splits both for closed-set and open-set settings, use MegaDescriptor - a foundational model for wildlife re-identification - for baseline performance and host a leaderboard with the best results. We publicly publish the dataset and the codes used to create it in the wildlife-datasets library, making WildlifeReID-10k both highly curated and easy to use.
WildlifeDatasets: An open-source toolkit for animal re-identification
Nov 15, 2023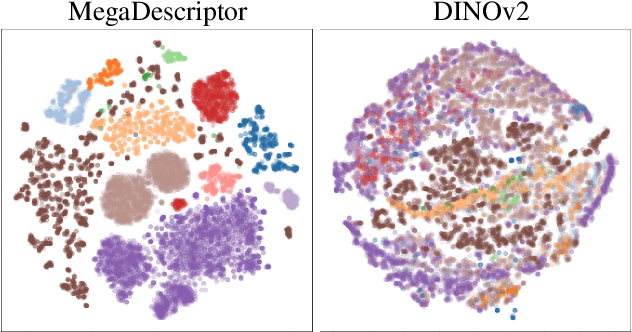


Abstract:In this paper, we present WildlifeDatasets (https://github.com/WildlifeDatasets/wildlife-datasets) - an open-source toolkit intended primarily for ecologists and computer-vision / machine-learning researchers. The WildlifeDatasets is written in Python, allows straightforward access to publicly available wildlife datasets, and provides a wide variety of methods for dataset pre-processing, performance analysis, and model fine-tuning. We showcase the toolkit in various scenarios and baseline experiments, including, to the best of our knowledge, the most comprehensive experimental comparison of datasets and methods for wildlife re-identification, including both local descriptors and deep learning approaches. Furthermore, we provide the first-ever foundation model for individual re-identification within a wide range of species - MegaDescriptor - that provides state-of-the-art performance on animal re-identification datasets and outperforms other pre-trained models such as CLIP and DINOv2 by a significant margin. To make the model available to the general public and to allow easy integration with any existing wildlife monitoring applications, we provide multiple MegaDescriptor flavors (i.e., Small, Medium, and Large) through the HuggingFace hub (https://huggingface.co/BVRA).
SeaTurtleID2022: A long-span dataset for reliable sea turtle re-identification
Nov 09, 2023



Abstract:This paper introduces the first public large-scale, long-span dataset with sea turtle photographs captured in the wild -- SeaTurtleID2022 (https://www.kaggle.com/datasets/wildlifedatasets/seaturtleid2022). The dataset contains 8729 photographs of 438 unique individuals collected within 13 years, making it the longest-spanned dataset for animal re-identification. All photographs include various annotations, e.g., identity, encounter timestamp, and body parts segmentation masks. Instead of standard "random" splits, the dataset allows for two realistic and ecologically motivated splits: (i) a time-aware closed-set with training, validation, and test data from different days/years, and (ii) a time-aware open-set with new unknown individuals in test and validation sets. We show that time-aware splits are essential for benchmarking re-identification methods, as random splits lead to performance overestimation. Furthermore, a baseline instance segmentation and re-identification performance over various body parts is provided. Finally, an end-to-end system for sea turtle re-identification is proposed and evaluated. The proposed system based on Hybrid Task Cascade for head instance segmentation and ArcFace-trained feature-extractor achieved an accuracy of 86.8%.
Unrolled three-operator splitting for parameter-map learning in Low Dose X-ray CT reconstruction
Apr 17, 2023

Abstract:We propose a method for fast and automatic estimation of spatially dependent regularization maps for total variation-based (TV) tomography reconstruction. The estimation is based on two distinct sub-networks, with the first sub-network estimating the regularization parameter-map from the input data while the second one unrolling T iterations of the Primal-Dual Three-Operator Splitting (PD3O) algorithm. The latter approximately solves the corresponding TV-minimization problem incorporating the previously estimated regularization parameter-map. The overall network is then trained end-to-end in a supervised learning fashion using pairs of clean-corrupted data but crucially without the need of having access to labels for the optimal regularization parameter-maps.
SeaTurtleID: A novel long-span dataset highlighting the importance of timestamps in wildlife re-identification
Nov 18, 2022



Abstract:This paper introduces SeaTurtleID, the first public large-scale, long-span dataset with sea turtle photographs captured in the wild. The dataset is suitable for benchmarking re-identification methods and evaluating several other computer vision tasks. The dataset consists of 7774 high-resolution photographs of 400 unique individuals collected within 12 years in 1081 encounters. Each photograph is accompanied by rich metadata, e.g., identity label, head segmentation mask, and encounter timestamp. The 12-year span of the dataset makes it the longest-spanned public wild animal dataset with timestamps. By exploiting this unique property, we show that timestamps are necessary for an unbiased evaluation of animal re-identification methods because they allow time-aware splits of the dataset into reference and query sets. We show that time-unaware splits can lead to performance overestimation of more than 100% compared to the time-aware splits for both feature- and CNN-based re-identification methods. We also argue that time-aware splits correspond to more realistic re-identification pipelines than the time-unaware ones. We recommend that animal re-identification methods should only be tested on datasets with timestamps using time-aware splits, and we encourage dataset curators to include such information in the associated metadata.
Optimization with learning-informed differential equation constraints and its applications
Aug 25, 2020
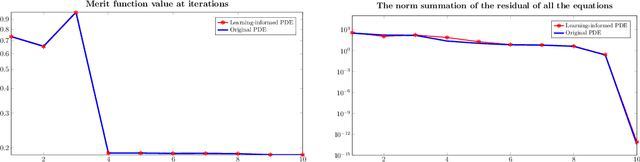


Abstract:Inspired by applications in optimal control of semilinear elliptic partial differential equations and physics-integrated imaging, differential equation constrained optimization problems with constituents that are only accessible through data-driven techniques are studied. A particular focus is on the analysis and on numerical methods for problems with machine-learned components. For a rather general context, an error analysis is provided, and particular properties resulting from artificial neural network based approximations are addressed. Moreover, for each of the two inspiring applications analytical details are presented and numerical results are provided.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge