Evangelos Papoutsellis
Why do we regularise in every iteration for imaging inverse problems?
Nov 01, 2024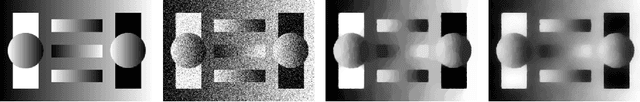

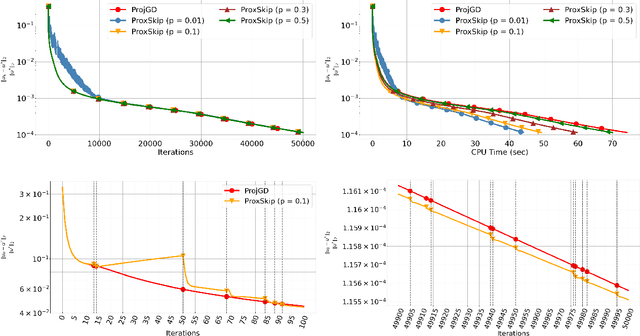
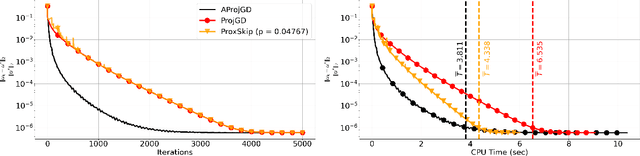
Abstract:Regularisation is commonly used in iterative methods for solving imaging inverse problems. Many algorithms involve the evaluation of the proximal operator of the regularisation term in every iteration, leading to a significant computational overhead since such evaluation can be costly. In this context, the ProxSkip algorithm, recently proposed for federated learning purposes, emerges as an solution. It randomly skips regularisation steps, reducing the computational time of an iterative algorithm without affecting its convergence. Here we explore for the first time the efficacy of ProxSkip to a variety of imaging inverse problems and we also propose a novel PDHGSkip version. Extensive numerical results highlight the potential of these methods to accelerate computations while maintaining high-quality reconstructions.
Stochastic Optimisation Framework using the Core Imaging Library and Synergistic Image Reconstruction Framework for PET Reconstruction
Jun 21, 2024Abstract:We introduce a stochastic framework into the open--source Core Imaging Library (CIL) which enables easy development of stochastic algorithms. Five such algorithms from the literature are developed, Stochastic Gradient Descent, Stochastic Average Gradient (-Am\'elior\'e), (Loopless) Stochastic Variance Reduced Gradient. We showcase the functionality of the framework with a comparative study against a deterministic algorithm on a simulated 2D PET dataset, with the use of the open-source Synergistic Image Reconstruction Framework. We observe that stochastic optimisation methods can converge in fewer passes of the data than a standard deterministic algorithm.
Unrolled three-operator splitting for parameter-map learning in Low Dose X-ray CT reconstruction
Apr 17, 2023

Abstract:We propose a method for fast and automatic estimation of spatially dependent regularization maps for total variation-based (TV) tomography reconstruction. The estimation is based on two distinct sub-networks, with the first sub-network estimating the regularization parameter-map from the input data while the second one unrolling T iterations of the Primal-Dual Three-Operator Splitting (PD3O) algorithm. The latter approximately solves the corresponding TV-minimization problem incorporating the previously estimated regularization parameter-map. The overall network is then trained end-to-end in a supervised learning fashion using pairs of clean-corrupted data but crucially without the need of having access to labels for the optimal regularization parameter-maps.
Enhanced hyperspectral tomography for bioimaging by spatiospectral reconstruction
Mar 08, 2021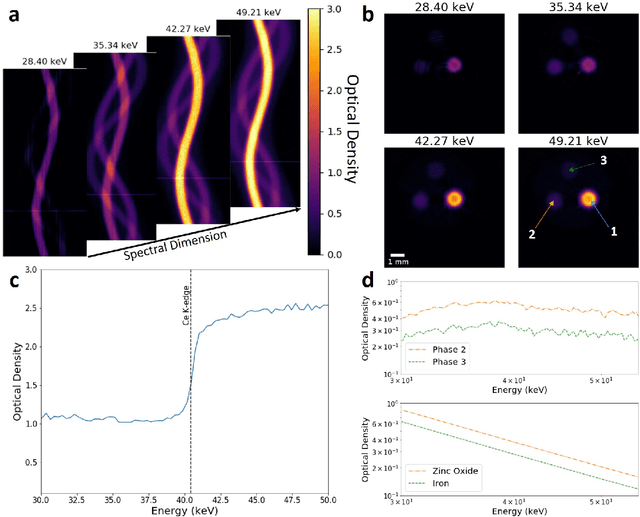
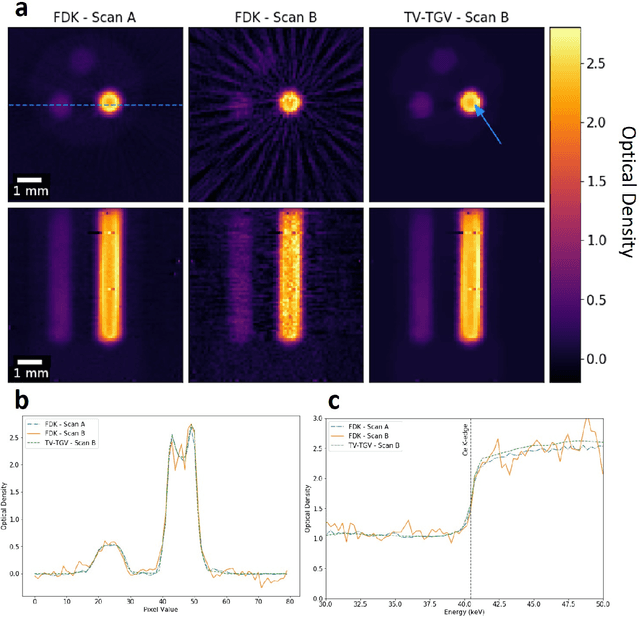
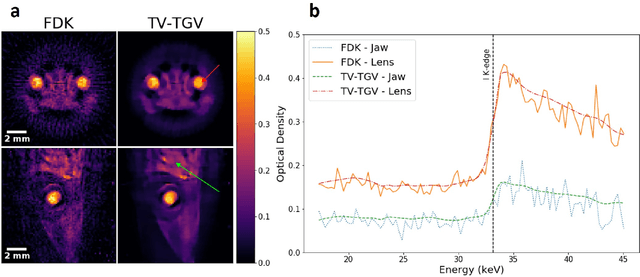
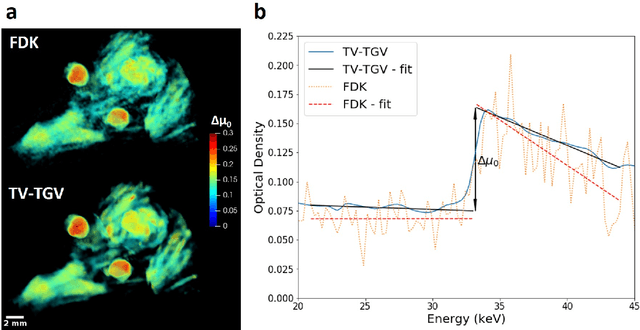
Abstract:Here we apply hyperspectral bright field imaging to collect computed tomographic images with excellent energy resolution (800 eV), applying it for the first time to map the distribution of stain in a fixed biological sample through its characteristic K-edge. Conventionally, because the photons detected at each pixel are distributed across as many as 200 energy channels, energy-selective images are characterised by low count-rates and poor signal-to-noise ratio. This means high X-ray exposures, long scan times and high doses are required to image unique spectral markers. Here, we achieve high quality energy-dispersive tomograms from low dose, noisy datasets using a dedicated iterative reconstruction algorithm. This exploits the spatial smoothness and inter-channel structural correlation in the spectral domain using two carefully chosen regularisation terms. For a multi-phase phantom, a reduction in scan time of 36 times is demonstrated. Spectral analysis methods including K-edge subtraction and absorption step-size fitting are evaluated for an ex vivo, single (iodine)-stained biological sample, where low chemical concentration and inhomogeneous distribution can affect soft tissue segmentation and visualisation. The reconstruction algorithms are available through the open-source Core Imaging Library. Taken together, these tools offer new capabilities for visualisation and elemental mapping, with promising applications for multiply-stained biological specimens.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge