Philip J. Withers
Digital Fingerprinting of Microstructures
Mar 25, 2022



Abstract:Finding efficient means of fingerprinting microstructural information is a critical step towards harnessing data-centric machine learning approaches. A statistical framework is systematically developed for compressed characterisation of a population of images, which includes some classical computer vision methods as special cases. The focus is on materials microstructure. The ultimate purpose is to rapidly fingerprint sample images in the context of various high-throughput design/make/test scenarios. This includes, but is not limited to, quantification of the disparity between microstructures for quality control, classifying microstructures, predicting materials properties from image data and identifying potential processing routes to engineer new materials with specific properties. Here, we consider microstructure classification and utilise the resulting features over a range of related machine learning tasks, namely supervised, semi-supervised, and unsupervised learning. The approach is applied to two distinct datasets to illustrate various aspects and some recommendations are made based on the findings. In particular, methods that leverage transfer learning with convolutional neural networks (CNNs), pretrained on the ImageNet dataset, are generally shown to outperform other methods. Additionally, dimensionality reduction of these CNN-based fingerprints is shown to have negligible impact on classification accuracy for the supervised learning approaches considered. In situations where there is a large dataset with only a handful of images labelled, graph-based label propagation to unlabelled data is shown to be favourable over discarding unlabelled data and performing supervised learning. In particular, label propagation by Poisson learning is shown to be highly effective at low label rates.
Enhanced hyperspectral tomography for bioimaging by spatiospectral reconstruction
Mar 08, 2021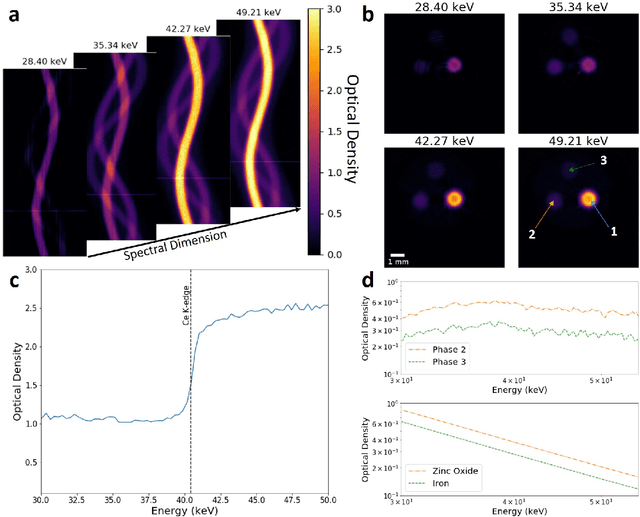
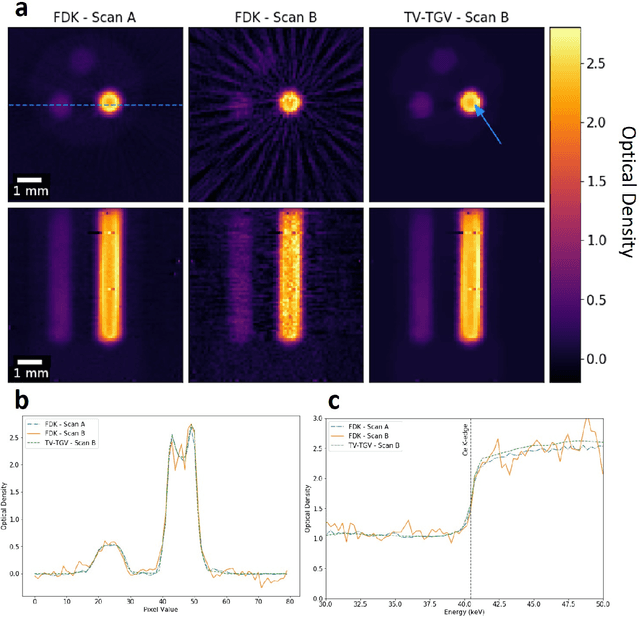
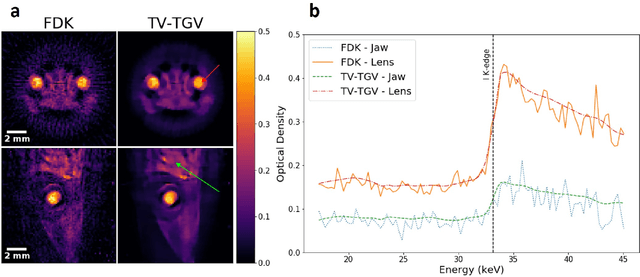
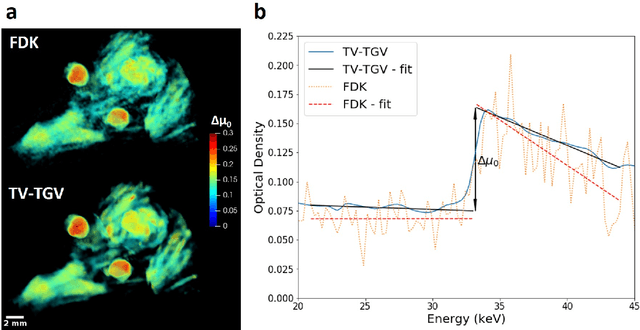
Abstract:Here we apply hyperspectral bright field imaging to collect computed tomographic images with excellent energy resolution (800 eV), applying it for the first time to map the distribution of stain in a fixed biological sample through its characteristic K-edge. Conventionally, because the photons detected at each pixel are distributed across as many as 200 energy channels, energy-selective images are characterised by low count-rates and poor signal-to-noise ratio. This means high X-ray exposures, long scan times and high doses are required to image unique spectral markers. Here, we achieve high quality energy-dispersive tomograms from low dose, noisy datasets using a dedicated iterative reconstruction algorithm. This exploits the spatial smoothness and inter-channel structural correlation in the spectral domain using two carefully chosen regularisation terms. For a multi-phase phantom, a reduction in scan time of 36 times is demonstrated. Spectral analysis methods including K-edge subtraction and absorption step-size fitting are evaluated for an ex vivo, single (iodine)-stained biological sample, where low chemical concentration and inhomogeneous distribution can affect soft tissue segmentation and visualisation. The reconstruction algorithms are available through the open-source Core Imaging Library. Taken together, these tools offer new capabilities for visualisation and elemental mapping, with promising applications for multiply-stained biological specimens.
Direct high-order edge-preserving regularization for tomographic image reconstruction
Sep 15, 2015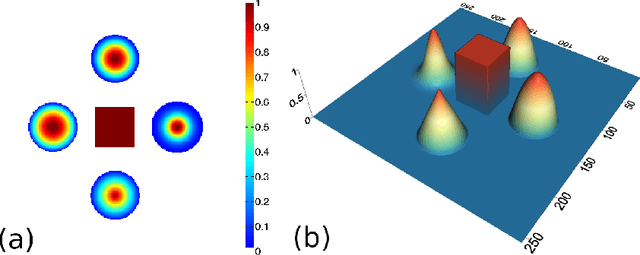
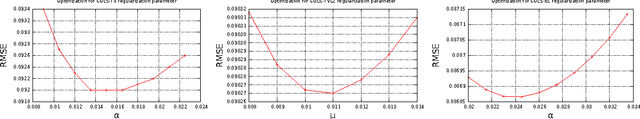
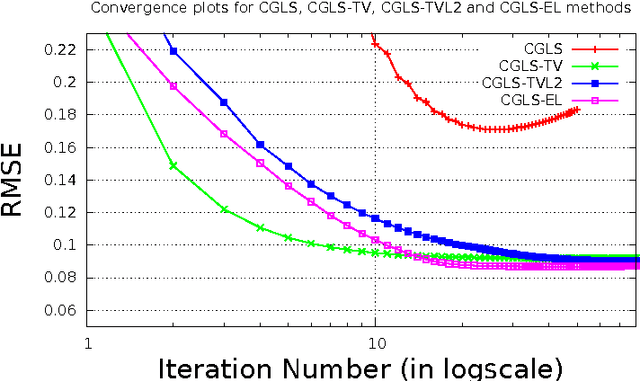
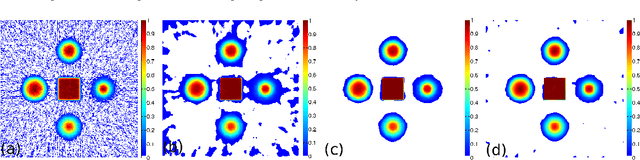
Abstract:In this paper we present a new two-level iterative algorithm for tomographic image reconstruction. The algorithm uses a regularization technique, which we call edge-preserving Laplacian, that preserves sharp edges between objects while damping spurious oscillations in the areas where the reconstructed image is smooth. Our numerical simulations demonstrate that the proposed method outperforms total variation (TV) regularization and it is competitive with the combined TV-L2 penalty. Obtained reconstructed images show increased signal-to-noise ratio and visually appealing structural features. Computer implementation and parameter control of the proposed technique is straightforward, which increases the feasibility of it across many tomographic applications. In this paper, we applied our method to the under-sampled computed tomography (CT) projection data and also considered a case of reconstruction in emission tomography The MATLAB code is provided to support obtained results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge